Hierarchy of sequence-dependent features associated with prokaryotic translation
- PMID: 14656971
- PMCID: PMC403808
- DOI: 10.1101/gr.1485203
Hierarchy of sequence-dependent features associated with prokaryotic translation
Abstract
Protein expression in the cell is affected by various sequence-dependent features. Several such sequence-dependent features have been individually studied,yet they have not been compared quantitatively in terms of their relative influence on protein expression,and a hierarchy of these elements has not been determined. Here we present a quantitative analysis examining sequence-dependent features involved in prokaryotic translation,namely,the base-pairing potential between the mRNA Shine-Dalgarno sequence and the ribosomal RNA,codon bias,and the identity of the stop codon. We analyzed these features both at intra- and intergenomic levels using the Escherichia coli and Haemophilus influenzae genomes. Within each genome,we examined the relationship between each feature and protein expression levels determined by 2D-gel analyses. At the intergenomic level,comparative genomic principles were applied to study the relative preservation of the different sequence-dependent properties between orthologs. From these analyses,we determined that biased codon usage is the property that is most highly associated with protein expression and that is most conserved. The identity of the stop codon and the base-pairing potential of the mRNA Shine-Dalgarno sequence and the rRNA seem to have less of an effect on protein expression.
Figures
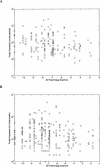

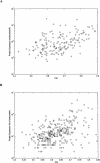
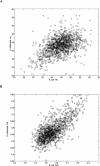
References
-
- Blattner, F.R., Plunkett III, G., Bloch, C.A., Perna, N.T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J.D., Rode, C.K., Mayhew, G.F., et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277: 1453-1474. - PubMed
WEB SITE REFERENCES
-
- http://www.ncbi.nlm.nih.gov; NCBI home page.
-
- http://opal.biology.gatech.edu/GeneMark/GeneMarkS/index.html; GeneMarkS gene predictions.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
