Discontinuous movement and conformational change during pausing and termination by T7 RNA polymerase
- PMID: 14657021
- PMCID: PMC291813
- DOI: 10.1093/emboj/cdg618
Discontinuous movement and conformational change during pausing and termination by T7 RNA polymerase
Abstract
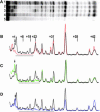
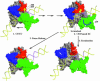
Time-resolved characterization of T7 RNA polymerase pausing and terminating at a class II termination site has been carried out using site-specifically tethered chemical nucleases. The data indicate that T7RNAP normally moves uniformly down the template as a rigid body. However, at the class II site this movement is interrupted, and the leading edge of the polymerase moves further along the DNA than the trailing edge. This discontinuous movement may persist until it can no longer be accommodated by conformational changes in the elongation complex, at which point the polymerase can either pause or terminate. Termination, but not pausing, is abrogated by introduction of a disulfide bond between the polymerase fingers and thumb subdomains. The introduced cysteines disrupt a thumb-fingers salt-bridge and, under reducing conditions, this mutant enzyme shows reduced processivity coincident with extension of the RNA to 5 nt. These observations suggest that termination requires that the thumb and fingers subdomains move apart, in a reversal of a conformational change important for initially forming a stable transcription complex.
Figures





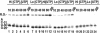
Similar articles
-
Probing conformational changes in T7 RNA polymerase during initiation and termination by using engineered disulfide linkages.Proc Natl Acad Sci U S A. 2005 Dec 6;102(49):17612-7. doi: 10.1073/pnas.0508865102. Epub 2005 Nov 21. Proc Natl Acad Sci U S A. 2005. PMID: 16301518 Free PMC article.
-
Pausing and termination by bacteriophage T7 RNA polymerase.J Mol Biol. 1998 Jul 10;280(2):201-13. doi: 10.1006/jmbi.1998.1854. J Mol Biol. 1998. PMID: 9654445
-
Mechanism of inhibition of bacteriophage T7 RNA polymerase by T7 lysozyme.J Mol Biol. 1997 May 30;269(1):10-27. doi: 10.1006/jmbi.1997.1016. J Mol Biol. 1997. PMID: 9192997
-
Transcription regulation, initiation, and "DNA scrunching" by T7 RNA polymerase.Cold Spring Harb Symp Quant Biol. 1998;63:263-7. doi: 10.1101/sqb.1998.63.263. Cold Spring Harb Symp Quant Biol. 1998. PMID: 10384290 Review. No abstract available.
-
Structure and function in promoter escape by T7 RNA polymerase.Prog Nucleic Acid Res Mol Biol. 2005;80:323-47. doi: 10.1016/S0079-6603(05)80008-X. Prog Nucleic Acid Res Mol Biol. 2005. PMID: 16164978 Review. No abstract available.
Cited by
-
Probing conformational changes in T7 RNA polymerase during initiation and termination by using engineered disulfide linkages.Proc Natl Acad Sci U S A. 2005 Dec 6;102(49):17612-7. doi: 10.1073/pnas.0508865102. Epub 2005 Nov 21. Proc Natl Acad Sci U S A. 2005. PMID: 16301518 Free PMC article.
-
Characterization of a novel RNA polymerase II arrest site which lacks a weak 3' RNA-DNA hybrid.Nucleic Acids Res. 2004 Mar 26;32(6):1904-16. doi: 10.1093/nar/gkh505. Print 2004. Nucleic Acids Res. 2004. PMID: 15047857 Free PMC article.
-
Involvement of the TPR2 subdomain movement in the activities of phi29 DNA polymerase.Nucleic Acids Res. 2009 Jan;37(1):193-203. doi: 10.1093/nar/gkn928. Epub 2008 Nov 25. Nucleic Acids Res. 2009. PMID: 19033368 Free PMC article.
-
Mechanism of T7 RNAP pausing and termination at the T7 concatemer junction: a local change in transcription bubble structure drives a large change in transcription complex architecture.J Mol Biol. 2008 Feb 15;376(2):541-53. doi: 10.1016/j.jmb.2007.11.090. Epub 2007 Dec 4. J Mol Biol. 2008. PMID: 18166198 Free PMC article.
-
Comparison of Pause Predictions of Two Sequence-Dependent Transcription Models.J Stat Mech. 2010 Dec;2010:P12007. doi: 10.1088/1742-5468/2010/12/P12007. J Stat Mech. 2010. PMID: 22446379 Free PMC article.
References
-
- Boudvillain M., Schwartz,A. and Rahmouni,A.R. (2002) Limited topological alteration of the T7 RNA polymerase active center at intrinsic termination site. Biochemistry, 4, 3137–3146. - PubMed
-
- Brieba L.G., Gopal,V. and Sousa,R. (2001) Scanning mutagenesis reveals roles for helix n of the bacteriophage T7 RNA polymerase thumb subdomain in transcription complex stability, pausing, and termination. J. Biol. Chem., 276, 10306–10313. - PubMed
-
- Cheetham G. and Steitz,T.A. (1999) Structure of a transcribing T7 RNA polymerase initiation complex. Science, 286, 2305–2309. - PubMed
-
- Greiner D.P., Miyake,R., Moran,J.K., Jones,A.D., Negishi,T., Ishihama,A. and Meares,C.F. (1997) Synthesis of the protein cutting reagent iron (S)-1-(p-bromoacetamidobenzyl)ethylenediaminetetra acetate and conjugation to cysteine side chains. Bioconjug. Chem., 8, 44–48. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

