Growth inhibition by the mammalian SWI-SNF subunit Brm is regulated by acetylation
- PMID: 14657023
- PMCID: PMC291816
- DOI: 10.1093/emboj/cdg621
Growth inhibition by the mammalian SWI-SNF subunit Brm is regulated by acetylation
Abstract
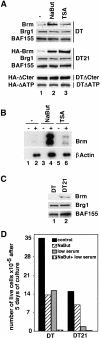
In mammalian cells, the SWI-SNF chromatin-remodeling complex is a regulator of cell proliferation, and overexpression of the catalytic subunit Brm interferes with cell cycle progression. Here, we show that treatment with histone deacetylase (HDAC) inhibitors reduces the inhibitory effect of Brm on the growth of mouse fibroblasts. This observation led to the identification of two carboxy-terminal acetylation sites in the Brm protein. Mutation of these sites into non-acetylatable sequences increased both the growth-inhibitory and the transcriptional activities of Brm. We also show that culture in the presence of HDAC inhibitors facilitates the isolation of clones overexpressing Brm. Removal of the HDAC inhibitors from the growth medium of these clones leads to downregulation of cyclin D1. This downregulation is absent in cell transformed by oncogenic ras.
Figures




References
-
- Abdollahi A., Pisarcik,D., Roberts,D., Weinstein,J., Cairns,P. and Hamilton,T.C. (2003) LOT1 (PLAGL1/ZAC1), the candidate tumor suppressor gene at chromosome 6q24–25, is epigenetically regulated in cancer. J. Biol. Chem., 278, 6041–6049. - PubMed
-
- Agalioti T., Chen,G. and Thanos,D. (2002) Deciphering the transcriptional histone acetylation code for a human gene. Cell, 111, 381–392. - PubMed
-
- Bannister A.J., Miska,E.A., Gorlich,D. and Kouzarides,T. (2000) Acetylation of importin-α nuclear import factors by CBP/p300. Curr. Biol., 10, 467–470. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

