Transcriptional activation of known and novel apoptotic pathways by Nur77 orphan steroid receptor
- PMID: 14657025
- PMCID: PMC291815
- DOI: 10.1093/emboj/cdg620
Transcriptional activation of known and novel apoptotic pathways by Nur77 orphan steroid receptor
Abstract
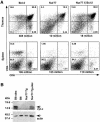
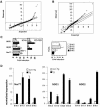
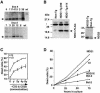
Nur77 is a nuclear orphan steroid receptor that has been implicated in negative selection. Expression of Nur77 in thymocytes and cell lines leads to apoptosis through a mechanism that remains unclear. In some cell lines, Nur77 was reported to act through a transcription-independent mechanism involving translocation to mitochondria, leading to cytochrome c release. However, we show here that Nur77-mediated apoptosis in thymocytes does not involve cytoplasmic cytochrome c release and cannot be rescued by Bcl-2. Microarray analysis shows that Nur77 induces many genes, including two novel genes (NDG1, NDG2) and known apoptotic genes FasL and TRAIL. Characterization of NDG1 and NDG2 indicates that NDG1 initiates a novel apoptotic pathway in a Bcl-2-independent manner. Thus Nur77-mediated apoptosis in T cells involves Bcl-2 independent transcriptional activation of several known and novel apoptotic pathways.
Figures

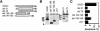

Similar articles
-
Functional redundancy of the Nur77 and Nor-1 orphan steroid receptors in T-cell apoptosis.EMBO J. 1997 Apr 15;16(8):1865-75. doi: 10.1093/emboj/16.8.1865. EMBO J. 1997. PMID: 9155013 Free PMC article.
-
Regulation of the Nur77 orphan steroid receptor in activation-induced apoptosis.Mol Cell Biol. 1995 Nov;15(11):6364-76. doi: 10.1128/MCB.15.11.6364. Mol Cell Biol. 1995. PMID: 7565789 Free PMC article.
-
Requirement for the orphan steroid receptor Nur77 in apoptosis of T-cell hybridomas.Nature. 1994 Jan 20;367(6460):277-81. doi: 10.1038/367277a0. Nature. 1994. PMID: 8121493
-
Molecular characterization of the Nur77 orphan steroid receptor in apoptosis.Int Arch Allergy Immunol. 1994 Dec;105(4):344-6. doi: 10.1159/000236780. Int Arch Allergy Immunol. 1994. PMID: 7981602 Review.
-
p53 and Nur77/TR3 - transcription factors that directly target mitochondria for cell death induction.Oncogene. 2006 Aug 7;25(34):4725-43. doi: 10.1038/sj.onc.1209601. Oncogene. 2006. PMID: 16892086 Review.
Cited by
-
Rethinking peripheral T cell tolerance: checkpoints across a T cell's journey.Nat Rev Immunol. 2021 Apr;21(4):257-267. doi: 10.1038/s41577-020-00454-2. Epub 2020 Oct 19. Nat Rev Immunol. 2021. PMID: 33077935 Review.
-
Regulation of vascular smooth muscle cell proliferation by nuclear orphan receptor Nur77.Mol Cell Biochem. 2010 Aug;341(1-2):159-66. doi: 10.1007/s11010-010-0447-0. Epub 2010 Apr 22. Mol Cell Biochem. 2010. PMID: 20411306
-
Microbiota Ecosystem in Recurrent Cystitis and the Immunological Microenvironment of Urothelium.Healthcare (Basel). 2023 Feb 10;11(4):525. doi: 10.3390/healthcare11040525. Healthcare (Basel). 2023. PMID: 36833059 Free PMC article. Review.
-
Minireview: Nuclear receptors, hematopoiesis, and stem cells.Mol Endocrinol. 2010 Jan;24(1):1-10. doi: 10.1210/me.2009-0332. Epub 2009 Nov 24. Mol Endocrinol. 2010. PMID: 19934345 Free PMC article. Review.
-
Non-redundant function of the MEK5-ERK5 pathway in thymocyte apoptosis.EMBO J. 2008 Jul 9;27(13):1896-906. doi: 10.1038/emboj.2008.114. Epub 2008 Jun 12. EMBO J. 2008. PMID: 18548009 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

