A La protein requirement for efficient pre-tRNA folding
- PMID: 14657028
- PMCID: PMC291820
- DOI: 10.1093/emboj/cdg625
A La protein requirement for efficient pre-tRNA folding
Abstract
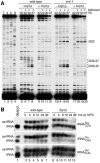
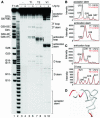
The La protein protects the 3' ends of many nascent small RNAs from exonucleases. Here we report that La is required for efficient folding of certain pre-tRNAs. A mutation in pre-tRNA(Arg)(CCG) causes yeast cells to be cold-sensitive and to require the La protein Lhp1p for efficient growth. When the mutant cells are grown at low temperature, or when Lhp1p is depleted, mature tRNA(Arg)(CCG) is not efficiently aminoacylated. The mutation causes the anticodon stem of pre-tRNA(Arg)(CCG) to misfold into an alternative helix in vitro. Intragenic suppressor mutations that disrupt the misfolded helix or strengthen the correct helix alleviate the requirement for Lhp1p, providing evidence that the anticodon stem misfolds in vivo. Chemical and enzymatic footprinting experiments suggest a model in which Lhp1p stabilizes the correctly folded stem. Lhp1p is also required for efficient aminoacylation of two wild-type tRNAs when yeast are grown at low temperature. These experiments reveal that pre-tRNAs can require protein assistance for efficient folding in vivo.
Figures





References
-
- Dammel C.S. and Noller,H.F. (1993) A cold-sensitive mutation in 16S rRNA provides evidence for helical switching in ribosome assembly. Genes Dev., 7, 660–670. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

