Nsl1p is essential for the establishment of bipolarity and the localization of the Dam-Duo complex
- PMID: 14657030
- PMCID: PMC291831
- DOI: 10.1093/emboj/cdg636
Nsl1p is essential for the establishment of bipolarity and the localization of the Dam-Duo complex
Abstract
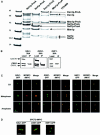
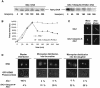
We identified a physical complex consisting of Mtw1p, an established kinetochore protein, with Nnf1p, Nsl1p and Dsn1p and have demonstrated that Nnf1p, Nsl1p and Dsn1p localize to the Saccharomyces cerevisiae kinetochore. When challenged prior to metaphase, the temperature-sensitive mutants nsl1-16 and nsl1-42 as well as Nsl1p-depleted cells failed to establish a bipolar spindle-kinetochore interaction and executed monopolar segregation of sister chromatids. In contrast, an nsl1-16 defect could not be evoked after the establishment of bipolarity. The observed phenotype is characteristic of that of mutants with defects in the protein kinase Ipl1p or components of the Dam-Duo kinetochore complex. However nsl1 mutants did not exhibit a defect in microtubule-kinetochore untethering as the ipl1-321 mutant does. Instead, they exhibited a severe defect in the kinetochore localization of the Dam-Duo complex suggesting this to be the cause for the failure of nsl1 cells to establish bipolarity. Moreover the analysis of Nsl1p-depleted cells indicated that Nsl1p is required for the spindle checkpoint and kinetochore integrity.
Figures





References
-
- Cheeseman I.M., Anderson,S., Jwa,M., Green,E.M., Kang,J., Yates,J.R.,3rd, Chan,C.S., Drubin,D.G. and Barnes,G. (2002a) Phospho-regulation of kinetochore–microtubule attachments by the Aurora kinase Ipl1p. Cell, 111, 163–172. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

