Dendritic cells initiate immune control of epstein-barr virus transformation of B lymphocytes in vitro
- PMID: 14657218
- PMCID: PMC2194129
- DOI: 10.1084/jem.20030646
Dendritic cells initiate immune control of epstein-barr virus transformation of B lymphocytes in vitro
Abstract
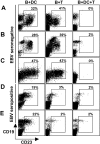
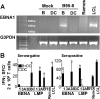
The initiation of cell-mediated immunity to Epstein-Barr virus (EBV) has been analyzed with cells from EBV-seronegative blood donors in culture. The addition of dendritic cells (DCs) is essential to prime naive T cells that recognize EBV-latent antigens in enzyme-linked immunospot assays for interferon gamma secretion and eradicate transformed B cells in regression assays. In contrast, DCs are not required to control the outgrowth of EBV-transformed B lymphocytes from seropositive donors. Enriched CD4+ and CD8+ T cells mediate regression of EBV-transformed cells in seronegative and seropositive donors, but the kinetics of T-dependent regression occurs with much greater speed with seropositives. EBV infection of DCs cannot be detected by reverse transcription-polymerase chain reaction with primers specific for mRNA for the EBNA1 U and K exons. Instead, DCs capture B cell debris and generate T cells specific for EBV latency antigens. We suggest that the cross-presentation of EBV-latent antigens from infected B cells by DCs is required for the initiation of EBV-specific immune control in vivo and that future EBV vaccine strategies should target viral antigens to DCs.
Figures





Similar articles
-
T cells specific for different latent and lytic viral proteins efficiently control Epstein-Barr virus-transformed B cells.Cytotherapy. 2015 Sep;17(9):1280-91. doi: 10.1016/j.jcyt.2015.06.003. Cytotherapy. 2015. PMID: 26276009
-
Dendritic cells cross-present latency gene products from Epstein-Barr virus-transformed B cells and expand tumor-reactive CD8(+) killer T cells.J Exp Med. 2001 Feb 5;193(3):405-11. doi: 10.1084/jem.193.3.405. J Exp Med. 2001. PMID: 11157061 Free PMC article.
-
Human CD4(+) T lymphocytes consistently respond to the latent Epstein-Barr virus nuclear antigen EBNA1.J Exp Med. 2000 May 15;191(10):1649-60. doi: 10.1084/jem.191.10.1649. J Exp Med. 2000. PMID: 10811859 Free PMC article.
-
Contrasting roles of dendritic cells and B cells in the immune control of Epstein-Barr virus.Curr Top Microbiol Immunol. 2003;276:55-76. doi: 10.1007/978-3-662-06508-2_3. Curr Top Microbiol Immunol. 2003. PMID: 12797443 Review.
-
Epstein-Barr virus evasion of CD8(+) and CD4(+) T cell immunity via concerted actions of multiple gene products.Semin Cancer Biol. 2008 Dec;18(6):397-408. doi: 10.1016/j.semcancer.2008.10.008. Epub 2008 Oct 25. Semin Cancer Biol. 2008. PMID: 18977445 Review.
Cited by
-
EBNA3B-deficient EBV promotes B cell lymphomagenesis in humanized mice and is found in human tumors.J Clin Invest. 2012 Apr;122(4):1487-502. doi: 10.1172/JCI58092. Epub 2012 Mar 12. J Clin Invest. 2012. PMID: 22406538 Free PMC article.
-
Editing antigen presentation: antigen transfer between human B lymphocytes and macrophages mediated by class A scavenger receptors.J Immunol. 2008 Sep 15;181(6):4043-51. doi: 10.4049/jimmunol.181.6.4043. J Immunol. 2008. PMID: 18768860 Free PMC article.
-
Immune escape by Epstein-Barr virus associated malignancies.Semin Cancer Biol. 2008 Dec;18(6):381-7. doi: 10.1016/j.semcancer.2008.10.002. Epub 2008 Oct 19. Semin Cancer Biol. 2008. PMID: 18996483 Free PMC article. Review.
-
Co-Infection and Cancer: Host-Pathogen Interaction between Dendritic Cells and HIV-1, HTLV-1, and Other Oncogenic Viruses.Viruses. 2022 Sep 14;14(9):2037. doi: 10.3390/v14092037. Viruses. 2022. PMID: 36146843 Free PMC article. Review.
-
Critical role of EBNA1-specific CD4+ T cells in the control of mouse Burkitt lymphoma in vivo.J Clin Invest. 2004 Aug;114(4):542-50. doi: 10.1172/JCI22053. J Clin Invest. 2004. PMID: 15314691 Free PMC article.
References
-
- Rickinson, A.B., and E. Kieff. 2001. Epstein-Barr virus. Fields Virology. P.M. Knipe and P.M. Howley, editors. Lippincott-Raven, Philadelphia. 2575–2627.
-
- Gahn, B., G. Hunt, C.M. Rooney, and H.E. Heslop. 2002. Immunotherapy to reconstitute immunity to DNA viruses. Semin. Hematol. 39:41–47. - PubMed
-
- Heslop, H.E., C.Y. Ng, C. Li, C.A. Smith, S.K. Loftin, R.A. Krance, M.K. Brenner, and C.M. Rooney. 1996. Long-term restoration of immunity against Epstein-Barr virus infection by adoptive transfer of gene-modified virus-specific T lymphocytes. Nat. Med. 2:551–555. - PubMed
-
- Boyle, G.J., M.G. Michaels, S.A. Webber, A.S. Knisely, G. Kurland, L.A. Cipriani, B.P. Griffith, and F.J. Fricker. 1997. Posttransplantation lymphoproliferative disorders in pediatric thoracic organ recipients. J. Pediatr. 131:309–313. - PubMed
-
- Metes, D., W. Storkus, A. Zeevi, K. Patterson, A. Logar, D. Rowe, M.A. Nalesnik, J.J. Fung, and A.S. Rao. 2000. Ex vivo generation of effective Epstein-Barr virus (EBV)-specific CD8+ cytotoxic T lymphocytes from the peripheral blood of immunocompetent Epstein Barr virus-seronegative individuals. Transplantation. 70:1507–1515. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

