Differentiation-induced colocalization of the KH-type splicing regulatory protein with polypyrimidine tract binding protein and the c-src pre-mRNA
- PMID: 14657238
- PMCID: PMC329392
- DOI: 10.1091/mbc.e03-09-0692
Differentiation-induced colocalization of the KH-type splicing regulatory protein with polypyrimidine tract binding protein and the c-src pre-mRNA
Abstract
We have examined the subcellular localization of the KH-type splicing regulatory protein (KSRP). KSRP is a multidomain RNA-binding protein implicated in a variety of cellular processes, including splicing in the nucleus and mRNA localization in the cytoplasm. We find that KSRP is primarily nuclear with a localization pattern that most closely resembles that of polypyrimidine tract binding protein (PTB). Colocalization experiments of KSRP with PTB in a mouse neuroblastoma cell line determined that both proteins are present in the perinucleolar compartment (PNC), as well as in other nuclear enrichments. In contrast, HeLa cells do not show prominent KSRP staining in the PNC, even though PTB labeling identified the PNC in these cells. Because both PTB and KSRP interact with the c-src transcript to affect N1 exon splicing, we examined the localization of the c-src pre-mRNA by fluorescence in situ hybridization. The src transcript is present in specific foci within the nucleus that are presumably sites of src transcription but are not generally perinucleolar. In normally cultured neuroblastoma cells, these src RNA foci contain PTB, but little KSRP. However, upon induced neuronal differentiation of these cells, KSRP occurs in the same foci with src RNA. PTB localization remains unaffected. This differentiation-induced localization of KSRP with src RNA correlates with an increase in src exon N1 inclusion. These results indicate that PTB and KSRP do indeed interact with the c-src transcript in vivo, and that these associations change with the differentiated state of the cell.
Figures
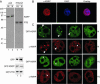
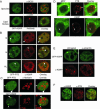


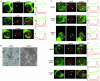
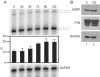
References
-
- Black, D.L. (1991). Does steric interference between splice sites block the splicing of a short c-src neuron-specific exon in non-neuronal cells? Genes Dev. 5, 389-402. - PubMed
-
- Black, D.L. (1992). Activation of c-src neuron-specific splicing by an unusual RNA element in vivo and in vitro. Cell 69, 795-807. - PubMed
-
- Carmo-Fonseca, M. (2002). The contribution of nuclear compartmentalization to gene regulation. Cell 108, 513-521. - PubMed
-
- Chen, C., Gherzi, R., Ong, S., Chan, E.L., Raijmakers, R., Pruijn, G.J.M., Stoeklin, G., Moroni, C., Mann, M., and Karin, M. (2001). AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107, 451-464. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

