WRN helicase and FEN-1 form a complex upon replication arrest and together process branchmigrating DNA structures associated with the replication fork
- PMID: 14657243
- PMCID: PMC329389
- DOI: 10.1091/mbc.e03-08-0567
WRN helicase and FEN-1 form a complex upon replication arrest and together process branchmigrating DNA structures associated with the replication fork
Abstract
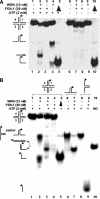
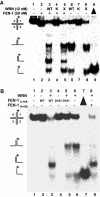
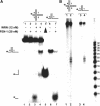
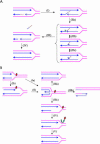
Werner Syndrome is a premature aging disorder characterized by genomic instability, elevated recombination, and replication defects. It has been hypothesized that defective processing of certain replication fork structures by WRN may contribute to genomic instability. Fluorescence resonance energy transfer (FRET) analyses show that WRN and Flap Endonuclease-1 (FEN-1) form a complex in vivo that colocalizes in foci associated with arrested replication forks. WRN effectively stimulates FEN-1 cleavage of branch-migrating double-flap structures that are the physiological substrates of FEN-1 during replication. Biochemical analyses demonstrate that WRN helicase unwinds the chicken-foot HJ intermediate associated with a regressed replication fork and stimulates FEN-1 to cleave the unwound product in a structure-dependent manner. These results provide evidence for an interaction between WRN and FEN-1 in vivo and suggest that these proteins function together to process DNA structures associated with the replication fork.
Figures








Similar articles
-
In vivo function of the conserved non-catalytic domain of Werner syndrome helicase in DNA replication.Hum Mol Genet. 2004 Oct 1;13(19):2247-61. doi: 10.1093/hmg/ddh234. Epub 2004 Jul 28. Hum Mol Genet. 2004. PMID: 15282207
-
Biochemical characterization of the WRN-FEN-1 functional interaction.Biochemistry. 2002 Oct 8;41(40):12204-16. doi: 10.1021/bi026031j. Biochemistry. 2002. PMID: 12356323
-
The interaction site of Flap Endonuclease-1 with WRN helicase suggests a coordination of WRN and PCNA.Nucleic Acids Res. 2005 Dec 2;33(21):6769-81. doi: 10.1093/nar/gki1002. Print 2005. Nucleic Acids Res. 2005. PMID: 16326861 Free PMC article.
-
Processing of DNA replication and repair intermediates by the concerted action of RecQ helicases and Rad2 structure-specific nucleases.Protein Pept Lett. 2008;15(1):89-102. doi: 10.2174/092986608783330369. Protein Pept Lett. 2008. PMID: 18221018 Review.
-
The Werner syndrome protein: linking the replication checkpoint response to genome stability.Aging (Albany NY). 2011 Mar;3(3):311-8. doi: 10.18632/aging.100293. Aging (Albany NY). 2011. PMID: 21389352 Free PMC article. Review.
Cited by
-
Biochemical Characterization of Middle East Respiratory Syndrome Coronavirus Helicase.mSphere. 2016 Sep 7;1(5):e00235-16. doi: 10.1128/mSphere.00235-16. eCollection 2016 Sep-Oct. mSphere. 2016. PMID: 27631026 Free PMC article.
-
A Link between Replicative Stress, Lamin Proteins, and Inflammation.Genes (Basel). 2021 Apr 9;12(4):552. doi: 10.3390/genes12040552. Genes (Basel). 2021. PMID: 33918867 Free PMC article. Review.
-
Human replication factor C stimulates flap endonuclease 1.J Biol Chem. 2009 Apr 17;284(16):10387-99. doi: 10.1074/jbc.M808893200. Epub 2009 Feb 9. J Biol Chem. 2009. PMID: 19208620 Free PMC article.
-
Interaction between the helicases genetically linked to Fanconi anemia group J and Bloom's syndrome.EMBO J. 2011 Feb 16;30(4):692-705. doi: 10.1038/emboj.2010.362. Epub 2011 Jan 14. EMBO J. 2011. PMID: 21240188 Free PMC article.
-
Control of structure-specific endonucleases to maintain genome stability.Nat Rev Mol Cell Biol. 2017 May;18(5):315-330. doi: 10.1038/nrm.2016.177. Epub 2017 Mar 22. Nat Rev Mol Cell Biol. 2017. PMID: 28327556 Review.
References
-
- Bambara, R.A., Murante, R.S., and Henricksen, L.A. (1997). Enzymes and reactions at the eukaryotic DNA replication fork. J. Biol. Chem. 272, 4647-4650. - PubMed
-
- Baynton, K., Otterlei, M., Bjoras, M., von Kobbe, C., Bohr, V.A., and Seeberg, E. (2003). WRN interacts physically and functionally with the recombination mediator protein RAD52. J. Biol. Chem. (Epub). - PubMed
-
- Brosh, R.M. Jr., and Bohr, V.A. (2002). Roles of the Werner syndrome protein in pathways required for maintenance of genome stability. Exp. Gerontol. 37, 491-506. - PubMed
-
- Brosh, R.M. Jr., Driscoll, H.C., Dianov, G.L., and Sommers, J.A. (2002a). Biochemical characterization of the WRN-FEN-1 functional interaction. Biochemistry 41, 12204-12216. - PubMed
-
- Brosh, R.M., Jr., Orren, D.K., Nehlin, J.O., Ravn, P.H., Kenny, M.K., Machwe, A., and Bohr, V.A. (1999). Functional and physical interaction between WRN helicase and human replication protein A. J. Biol. Chem. 274, 18341-18350. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

