NMR-detected hydrogen exchange and molecular dynamics simulations provide structural insight into fibril formation of prion protein fragment 106-126
- PMID: 14657385
- PMCID: PMC299804
- DOI: 10.1073/pnas.2433563100
NMR-detected hydrogen exchange and molecular dynamics simulations provide structural insight into fibril formation of prion protein fragment 106-126
Abstract
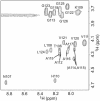
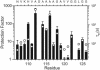
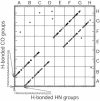
PrP106-126, a peptide corresponding to residues 107-127 of the human prion protein, induces neuronal cell death by apoptosis and causes proliferation and hypertrophy of glia, reproducing the main neuropathological features of prion-related transmissible spongiform encephalopathies, such as bovine spongiform encephalopathy and Creutzfeldt-Jakob disease. Although PrP106-126 has been shown to form amyloid-like fibrils in vitro, their structural properties have not been elucidated. Here, we investigate the conformational characteristics of a fibril-forming fragment of the mouse prion protein, MoPrP106-126, by using electron microscopy, CD spectroscopy, NMR-detected hydrogen-deuterium exchange measurements, and molecular dynamics simulations. The fibrils contain approximately 50% beta-sheet structure, and strong amide exchange protection is limited to the central portion of the peptide spanning the palindromic sequence VAGAAAAGAV. Molecular dynamics simulations indicate that MoPrP106-126 in water assumes a stable structure consisting of two four-stranded parallel beta-sheets that are tightly packed against each other by methyl-methyl interactions. Fibril formation involving polyalanine stacking is consistent with the experimental observations.
Figures


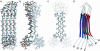
Similar articles
-
Dissection of conformational conversion events during prion amyloid fibril formation using hydrogen exchange and mass spectrometry.J Mol Biol. 2013 Sep 23;425(18):3510-21. doi: 10.1016/j.jmb.2013.06.009. Epub 2013 Jun 25. J Mol Biol. 2013. PMID: 23811055
-
Beta-sheet core of human prion protein amyloid fibrils as determined by hydrogen/deuterium exchange.Proc Natl Acad Sci U S A. 2007 Jan 30;104(5):1510-5. doi: 10.1073/pnas.0608447104. Epub 2007 Jan 22. Proc Natl Acad Sci U S A. 2007. PMID: 17242357 Free PMC article.
-
Conformational polymorphism of the PrP106-126 peptide in different environments: a molecular dynamics study.J Phys Chem B. 2006 Jan 26;110(3):1423-8. doi: 10.1021/jp052722o. J Phys Chem B. 2006. PMID: 16471693
-
Towards modeling of amyloid fibril structures.Front Biosci. 2008 May 1;13:4039-50. doi: 10.2741/2992. Front Biosci. 2008. PMID: 18508498 Review.
-
Simulations and computational analyses of prion protein conformations.Adv Protein Chem. 2001;57:107-37. doi: 10.1016/s0065-3233(01)57020-5. Adv Protein Chem. 2001. PMID: 11447688 Review. No abstract available.
Cited by
-
Interactions between the conserved hydrophobic region of the prion protein and dodecylphosphocholine micelles.J Biol Chem. 2012 Jan 13;287(3):1915-22. doi: 10.1074/jbc.M111.279364. Epub 2011 Nov 29. J Biol Chem. 2012. PMID: 22128151 Free PMC article.
-
The use of spin desalting columns in DMSO-quenched H/D-exchange NMR experiments.Protein Sci. 2013 Apr;22(4):486-91. doi: 10.1002/pro.2221. Epub 2013 Feb 11. Protein Sci. 2013. PMID: 23339068 Free PMC article.
-
N-terminal Prion Protein Peptides (PrP(120-144)) Form Parallel In-register β-Sheets via Multiple Nucleation-dependent Pathways.J Biol Chem. 2016 Oct 14;291(42):22093-22105. doi: 10.1074/jbc.M116.744573. Epub 2016 Aug 30. J Biol Chem. 2016. PMID: 27576687 Free PMC article.
-
Structural diversity and initial oligomerization of PrP106-126 studied by replica-exchange and conventional molecular dynamics simulations.PLoS One. 2014 Feb 19;9(2):e87266. doi: 10.1371/journal.pone.0087266. eCollection 2014. PLoS One. 2014. PMID: 24586266 Free PMC article.
-
How do amino acid substitutions affect the amyloidogenic properties and seeding efficiency of prion peptides.Amino Acids. 2013 Oct;45(4):785-96. doi: 10.1007/s00726-013-1522-0. Epub 2013 Jun 5. Amino Acids. 2013. PMID: 23736988 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

