NMR-detected hydrogen exchange and molecular dynamics simulations provide structural insight into fibril formation of prion protein fragment 106-126
- PMID: 14657385
- PMCID: PMC299804
- DOI: 10.1073/pnas.2433563100
NMR-detected hydrogen exchange and molecular dynamics simulations provide structural insight into fibril formation of prion protein fragment 106-126
Abstract
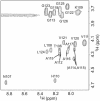
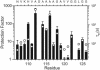
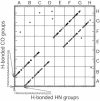
PrP106-126, a peptide corresponding to residues 107-127 of the human prion protein, induces neuronal cell death by apoptosis and causes proliferation and hypertrophy of glia, reproducing the main neuropathological features of prion-related transmissible spongiform encephalopathies, such as bovine spongiform encephalopathy and Creutzfeldt-Jakob disease. Although PrP106-126 has been shown to form amyloid-like fibrils in vitro, their structural properties have not been elucidated. Here, we investigate the conformational characteristics of a fibril-forming fragment of the mouse prion protein, MoPrP106-126, by using electron microscopy, CD spectroscopy, NMR-detected hydrogen-deuterium exchange measurements, and molecular dynamics simulations. The fibrils contain approximately 50% beta-sheet structure, and strong amide exchange protection is limited to the central portion of the peptide spanning the palindromic sequence VAGAAAAGAV. Molecular dynamics simulations indicate that MoPrP106-126 in water assumes a stable structure consisting of two four-stranded parallel beta-sheets that are tightly packed against each other by methyl-methyl interactions. Fibril formation involving polyalanine stacking is consistent with the experimental observations.
Figures


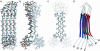
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

