Kinesin moves by an asymmetric hand-over-hand mechanism
- PMID: 14657506
- PMCID: PMC1523256
- DOI: 10.1126/science.1092985
Kinesin moves by an asymmetric hand-over-hand mechanism
Abstract
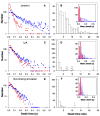
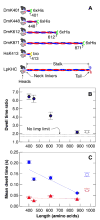
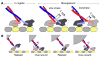
Kinesin is a double-headed motor protein that moves along microtubules in 8-nanometer steps. Two broad classes of model have been invoked to explain kinesin movement: hand-over-hand and inchworm. In hand-over-hand models, the heads exchange leading and trailing roles with every step, whereas no such exchange is postulated for inchworm models, where one head always leads. By measuring the stepwise motion of individual enzymes, we find that some kinesin molecules exhibit a marked alternation in the dwell times between sequential steps, causing these motors to "limp" along the microtubule. Limping implies that kinesin molecules strictly alternate between two different conformations as they step, indicative of an asymmetric, hand-over-hand mechanism.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

