Multiclass classification of microarray data with repeated measurements: application to cancer
- PMID: 14659020
- PMCID: PMC329422
- DOI: 10.1186/gb-2003-4-12-r83
Multiclass classification of microarray data with repeated measurements: application to cancer
Erratum in
- Genome Biol. 2005;6(13):405-405.4
Abstract
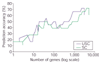
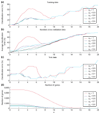
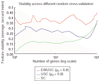
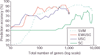
Prediction of the diagnostic category of a tissue sample from its gene-expression profile and selection of relevant genes for class prediction have important applications in cancer research. We have developed the uncorrelated shrunken centroid (USC) and error-weighted, uncorrelated shrunken centroid (EWUSC) algorithms that are applicable to microarray data with any number of classes. We show that removing highly correlated genes typically improves classification results using a small set of genes.
Figures

Similar articles
-
New gene selection method for multiclass tumor classification by class centroid.J Biomed Inform. 2009 Feb;42(1):59-65. doi: 10.1016/j.jbi.2008.05.011. Epub 2008 Jun 17. J Biomed Inform. 2009. PMID: 18835752
-
Tumor classification and marker gene prediction by feature selection and fuzzy c-means clustering using microarray data.BMC Bioinformatics. 2003 Dec 2;4:60. doi: 10.1186/1471-2105-4-60. BMC Bioinformatics. 2003. PMID: 14651757 Free PMC article.
-
Automated Detection of Cancer Associated Genes Using a Combined Fuzzy-Rough-Set-Based F-Information and Water Swirl Algorithm of Human Gene Expression Data.PLoS One. 2016 Dec 9;11(12):e0167504. doi: 10.1371/journal.pone.0167504. eCollection 2016. PLoS One. 2016. PMID: 27936033 Free PMC article.
-
Key aspects of analyzing microarray gene-expression data.Pharmacogenomics. 2007 May;8(5):473-82. doi: 10.2217/14622416.8.5.473. Pharmacogenomics. 2007. PMID: 17465711 Review.
-
Predictive ability of DNA microarrays for cancer outcomes and correlates: an empirical assessment.Lancet. 2003 Nov 1;362(9394):1439-44. doi: 10.1016/S0140-6736(03)14686-7. Lancet. 2003. PMID: 14602436 Review.
Cited by
-
Establishing Novel Molecular Subtypes of Appendiceal Cancer.Ann Surg Oncol. 2022 Mar;29(3):2118-2125. doi: 10.1245/s10434-021-10945-8. Epub 2021 Oct 30. Ann Surg Oncol. 2022. PMID: 34718915
-
Developing a Predictive Gene Classifier for Autism Spectrum Disorders Based upon Differential Gene Expression Profiles of Phenotypic Subgroups.N Am J Med Sci (Boston). 2013;6(3):10.7156/najms.2013.0603107. doi: 10.7156/najms.2013.0603107. N Am J Med Sci (Boston). 2013. PMID: 24363828 Free PMC article.
-
Gene selection and classification of microarray data using random forest.BMC Bioinformatics. 2006 Jan 6;7:3. doi: 10.1186/1471-2105-7-3. BMC Bioinformatics. 2006. PMID: 16398926 Free PMC article.
-
Optimized between-group classification: a new jackknife-based gene selection procedure for genome-wide expression data.BMC Bioinformatics. 2005 Sep 28;6:239. doi: 10.1186/1471-2105-6-239. BMC Bioinformatics. 2005. PMID: 16191195 Free PMC article.
-
A Dual Level Analysis with Evolutionary Computing and Swarm Models for Classification of Leukemia.Biomed Res Int. 2022 May 26;2022:2052061. doi: 10.1155/2022/2052061. eCollection 2022. Biomed Res Int. 2022. PMID: 35663047 Free PMC article.
References
-
- Lennon GG, Lehrach H. Hybridization analyses of arrayed cDNA libraries. Trends Genet. 1991;7:314–317. - PubMed
-
- Pietu G, Alibert O, Guichard V, Lamy B, Bois F, Leroy E, Mariage-Sampson R, Houlgatte R, Soularue P, Auffray C. Novel gene transcripts preferentially expressed in human muscles revealed by quantitative hybridization of a high density cDNA array. Genome Res. 1996;6:492–503. - PubMed
-
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. - PubMed
-
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

