Experimental examination of bacteriophage latent-period evolution as a response to bacterial availability
- PMID: 14660403
- PMCID: PMC310036
- DOI: 10.1128/AEM.69.12.7499-7506.2003
Experimental examination of bacteriophage latent-period evolution as a response to bacterial availability
Abstract
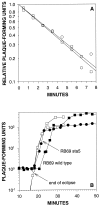
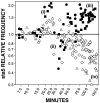
For obligately lytic bacteriophage (phage) a trade-off exists between fecundity (burst size) and latent period (a component of generation time). This trade-off occurs because release of phage progeny from infected bacteria coincides with destruction of the machinery necessary to produce more phage progeny. Here we employ phage mutants to explore issues of phage latent-period evolution as a function of the density of phage-susceptible bacteria. Theory suggests that higher bacterial densities should select for shorter phage latent periods. Consistently, we have found that higher host densities (>/== approximately 10(7) bacteria/ml) can enrich stocks of phage RB69 for variants that display shorter latent periods than the wild type. One such variant, dubbed sta5, displays a latent period that is approximately 70 to 80% of that of the wild type-which is nearly as short as the RB69 eclipse period-and which has a corresponding burst size that is approximately 30% of that of the wild type. We show that at higher host densities (>/== approximately 10(7) bacteria/ml) the sta5 phage can outcompete the RB69 wild type, though only under conditions of direct (same-culture) competition. We interpret this advantage as corresponding to slightly faster sta5 population growth, resulting in multifold increases in mutant frequency during same-culture growth. The sta5 advantage is lost, however, given indirect (different-culture) competition between the wild type and mutant or given same-culture competition but at lower densities of phage-susceptible bacteria (</= approximately 10(6) bacteria/ml). From these observations we suggest that phage displaying very short latent periods may be viewed as specialists for propagation when bacteria within cultures are highly prevalent and transmission between cultures is easily accomplished.
Figures




References
-
- Abedon, S. T. 1989. Selection for bacteriophage latent period length by bacterial density: a theoretical examination. Microb. Ecol. 18:79-88. - PubMed
-
- Abedon, S. T. 1990. Selection for lysis inhibition in bacteriophage. J. Theor. Biol. 146:501-511. - PubMed
-
- Abedon, S. T. 1994. Lysis and the interaction between free phages and infected cells, p. 397-405. In J. D. Karam et al. (ed.), Molecular biology of bacteriophage T4. ASM Press, Washington, D.C.
-
- Abedon, S. T. 1999. Bacteriophage T4 resistance to lysis-inhibition collapse. Genet. Res. 74:1-11. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

