Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast
- PMID: 14663140
- PMCID: PMC307643
- DOI: 10.1073/pnas.2632890100
Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast
Abstract
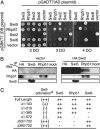
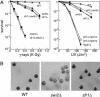
Homologous recombination is an important biological process that occurs in all organisms and facilitates genome rearrangements and repair of DNA double-strand breaks. Eukaryotic Rad51 proteins (Rad51sp or Rhp51 in fission yeast) are functional and structural homologs of bacterial RecA protein, an evolutionarily conserved protein that plays a key role in homologous pairing and strand exchange between homologous DNA molecules in vitro. Here we show that the fission yeast swi5+ gene, which was originally identified as a gene required for normal mating-type switching, encodes a protein conserved among eukaryotes and is involved in a previously uncharacterized Rhp51 (Rad51sp)-dependent recombination repair pathway that does not require the Rhp55/57 (Rad55/57sp) function. Protein interactions with both Swi5 and Rhp51 were found to be mediated by a domain common to Swi2 and Sfr1 (Swi five-dependent recombination repair protein 1, a previously uncharacterized protein with sequence similarity to the C-terminal part of Swi2). Genetic epistasis analyses suggest that the Swi5-Sfr1-Rhp51 interactions function specifically in DNA recombination repair, whereas the Swi5-Swi2-Rhp51 interactions may function, together with chromodomain protein Swi6 (HP1 homolog), in mating-type switching.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

