RNA interference: biology, mechanism, and applications
- PMID: 14665679
- PMCID: PMC309050
- DOI: 10.1128/MMBR.67.4.657-685.2003
RNA interference: biology, mechanism, and applications
Abstract
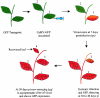
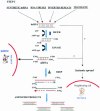
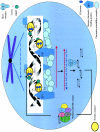
Double-stranded RNA-mediated interference (RNAi) is a simple and rapid method of silencing gene expression in a range of organisms. The silencing of a gene is a consequence of degradation of RNA into short RNAs that activate ribonucleases to target homologous mRNA. The resulting phenotypes either are identical to those of genetic null mutants or resemble an allelic series of mutants. Specific gene silencing has been shown to be related to two ancient processes, cosuppression in plants and quelling in fungi, and has also been associated with regulatory processes such as transposon silencing, antiviral defense mechanisms, gene regulation, and chromosomal modification. Extensive genetic and biochemical analysis revealed a two-step mechanism of RNAi-induced gene silencing. The first step involves degradation of dsRNA into small interfering RNAs (siRNAs), 21 to 25 nucleotides long, by an RNase III-like activity. In the second step, the siRNAs join an RNase complex, RISC (RNA-induced silencing complex), which acts on the cognate mRNA and degrades it. Several key components such as Dicer, RNA-dependent RNA polymerase, helicases, and dsRNA endonucleases have been identified in different organisms for their roles in RNAi. Some of these components also control the development of many organisms by processing many noncoding RNAs, called micro-RNAs. The biogenesis and function of micro-RNAs resemble RNAi activities to a large extent. Recent studies indicate that in the context of RNAi, the genome also undergoes alterations in the form of DNA methylation, heterochromatin formation, and programmed DNA elimination. As a result of these changes, the silencing effect of gene functions is exercised as tightly as possible. Because of its exquisite specificity and efficiency, RNAi is being considered as an important tool not only for functional genomics, but also for gene-specific therapeutic activities that target the mRNAs of disease-related genes.
Figures


References
-
- Aboobaker, A. A., and M. L. Blaxter. 2003. Use of RNA interference to investigate gene function in the human filarial nematode parasite Brugia malayi. Mol. Biochem. Parasitol. 129:41-51. - PubMed
-
- Abraham, N., D. J. Stojdl, P. L. Duncan, N. Methot, T. Ishii, M. Dube, B. C. Vanderhyden, H. L. Atkins, D. A. Gray, and M. W. McBurney. 1999. Characterization of transgenic mice with targeted disruption of the catalytic domain of the double-stranded RNA-dependent protein kinase, PKR. J. Biol. Chem. 274:5953-5962. - PubMed
-
- Akhtar, A., D. Zinc, and P. B. Becker. 2000. Chromodomains are protein-RNA interaction modules. Nature 407:405. - PubMed
-
- Al-Anouti, F., T. Quach, and S. Ananvoranich. 2003. Double-stranded RNA can mediate the suppression of uracil phosphoribosyltransferase expression in Toxoplasma gondii. Biochem. Biophys. Res. Commun. 302:316-323. - PubMed
-
- Al-Kaff, N. S., S. N. Covey, M. M. Kreike, A. M. Page, R. Pinder, and P. Dale. 1998. Transcriptional and posttranscriptional plant gene silencing in response to a pathogen. Science 279:2113-2115. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

