Role of viruses in human evolution
- PMID: 14666532
- PMCID: PMC7159740
- DOI: 10.1002/ajpa.10384
Role of viruses in human evolution
Abstract
The study of viral molecular genetics has produced a considerable body of research into the sequences and phylogenetic relationships of human and animal viruses. A review of this literature suggests that humans have been afflicted by viruses throughout their evolutionary history, although the number and types have changed. Some viruses show evidence of long-standing intimate relationship and cospeciation with hominids, while others are more recently acquired from other species, including African monkeys and apes while our line was evolving in that continent, and domesticated animals and rodents since the Neolithic. Viral selection for specific resistance polymorphisms is unlikely, but in conjunction with other parasites, viruses have probably contributed to selection pressure maintaining major histocompatibility complex (MHC) diversity and a strong immune response. They may also have played a role in the loss in our lineage of N-glycolylneuraminic acid (Neu5Gc), a cell-surface receptor for many infectious agents. Shared viruses could have affected hominid species diversity both by promoting divergence and by weeding out less resistant host populations, while viruses carried by humans and other animals migrating out of Africa may have contributed to declines in other populations. Endogenous retroviral insertions since the divergence between humans and chimpanzees were capable of directly affecting hominid evolution through changes in gene expression and development.
Copyright 2003 Wiley-Liss, Inc.
Figures
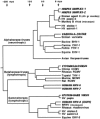


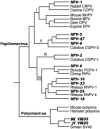
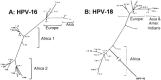
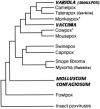
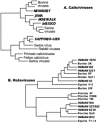
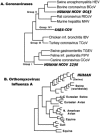

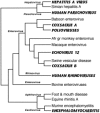
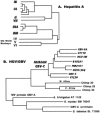

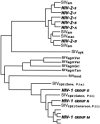
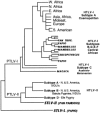

References
-
- Adams NJ, Prescott LE, Jarvis LM, Lewis JCM, McClure MO, Smith DB, Simmonds P. 1998. Detection in chimpanzees of a novel flavivirus related to GB virus‐C/hepatitis G virus. J Gen Virol 79: 1871–1877. - PubMed
-
- Akopov SB, Nikolaev LG, Khil PP, Lebedev YB, Sverdlov ED. 1998. Long terminal repeats of human endogenous retrovirus K family (HERV‐K) specifically bind host cell nuclear proteins. FEBS Lett 421: 229–233. - PubMed
-
- Amengual B, Whitby JE, King A, Cobo JS, Bourhy H. 1997. Evolution of European bat lyssaviruses. J Gen Virol 78: 2319–2328. - PubMed
-
- Armelagos GJ. 1990. Health and disease in prehistoric populations in transition In: Swedlund AC, Armelagos GJ, editors. Disease in populations in transition. New York: Bergin & Garvey; p 127–144.
-
- Armelagos GJ, Dewey JR. 1978. Evolutionary response to human infectious diseases In: Logan MB, Hunt EE, Sr, editors. Health and the human condition: perspectives in medical anthropology. Belmont, CA: Wadsworth Publishing Co. p 101–107.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

