Global gene expression responses of fission yeast to ionizing radiation
- PMID: 14668484
- PMCID: PMC329398
- DOI: 10.1091/mbc.e03-08-0569
Global gene expression responses of fission yeast to ionizing radiation
Abstract
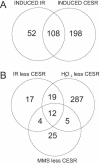
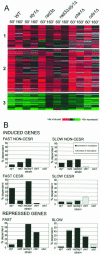
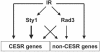
A coordinated transcriptional response to DNA-damaging agents is required to maintain genome stability. We have examined the global gene expression responses of the fission yeast Schizosaccharomyces pombe to ionizing radiation (IR) by using DNA microarrays. We identified approximately 200 genes whose transcript levels were significantly altered at least twofold in response to 500 Gy of gamma IR in a temporally defined manner. The majority of induced genes were core environmental stress response genes, whereas the remaining genes define a transcriptional response to DNA damage in fission yeast. Surprisingly, few DNA repair and checkpoint genes were transcriptionally modulated in response to IR. We define a role for the stress-activated mitogen-activated protein kinase Sty1/Spc1 and the DNA damage checkpoint kinase Rad3 in regulating core environmental stress response genes and IR-specific response genes, both independently and in concert. These findings suggest a complex network of regulatory pathways coordinate gene expression responses to IR in eukaryotes.
Figures

References
-
- Allen, J.B., Zhou, Z., Siede, W., Friedberg, E.C., and Elledge, S.J. (1994). The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 8, 2401-2415. - PubMed
-
- Bang, D.D., Ketting, R., de Ruijter, M., Brandsma, J.A., Verhage, R.A., van de Putte, P., and Brouwer, J. (1996). Cloning of Schizosaccharomyces pombe rph16+, a gene homologous to the Saccharomyces cerevisiae RAD16 gene. Mutat. Res. 364, 57-71. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

