Chlamydomonas reinhardtii secretes compounds that mimic bacterial signals and interfere with quorum sensing regulation in bacteria
- PMID: 14671013
- PMCID: PMC316294
- DOI: 10.1104/pp.103.029918
Chlamydomonas reinhardtii secretes compounds that mimic bacterial signals and interfere with quorum sensing regulation in bacteria
Abstract
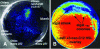
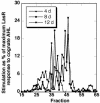
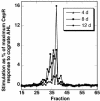
The unicellular soil-freshwater alga Chlamydomonas reinhardtii was found to secrete substances that mimic the activity of the N-acyl-L-homoserine lactone (AHL) signal molecules used by many bacteria for quorum sensing regulation of gene expression. More than a dozen chemically separable but unidentified substances capable of specifically stimulating the LasR or CepR but not the LuxR, AhyR, or CviR AHL bacterial quorum sensing reporter strains were detected in ethyl acetate extracts of C. reinhardtii culture filtrates. Colonies of C. reinhardtii and Chlorella spp. stimulated quorum sensing-dependent luminescence in Vibrio harveyi, indicating that these algae may produce compounds that affect the AI-2 furanosyl borate diester-mediated quorum sensing system of Vibrio spp. Treatment of the soil bacterium Sinorhizobium meliloti with a partially purified LasR mimic from C. reinhardtii affected the accumulation of 16 of the 25 proteins that were altered in response to the bacterium's own AHL signals, providing evidence that the algal mimic affected quorum sensing-regulated functions in this wild-type bacterium. Peptide mass fingerprinting identified 32 proteins affected by the bacterium's AHLs or the purified algal mimic, including GroEL chaperonins, the nitrogen regulatory protein PII, and a GTP-binding protein. The algal mimic was able to cancel the stimulatory effects of bacterial AHLs on the accumulation of seven of these proteins, providing evidence that the secretion of AHL mimics by the alga could be effective in disruption of quorum sensing in naturally encountered bacteria.
Figures

References
-
- Arcondeguy T, Huez I, Tillard P, Gangneux C, de Billy F, Gojon A, Truchet G, Kahn D (1997) The Rhizobium meliloti PII protein, which controls bacterial nitrogen metabolism, affects alfalfa nodule development. Genes Dev 11: 1194-1206 - PubMed
-
- Bassler BL, Wright M, Showalter RE, Silverman MR (1993) Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol 9: 773-786 - PubMed
-
- Bauer WD, Teplitski M (2001) Can plants manipulate bacterial quorum sensing? Aust J Plant Physiol 28: 913-921
-
- Chen H, Higgins J, Kondorosi E, Kondorosi A, Djordjevic MA, Weinman JJ, Rolfe BG (2000a) Identification of nolR-regulated proteins in Sinorhizobium meliloti using proteome analysis. Electrophoresis 21: 3823-3832 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

