Residues of human cytomegalovirus DNA polymerase catalytic subunit UL54 that are necessary and sufficient for interaction with the accessory protein UL44
- PMID: 14671097
- PMCID: PMC303418
- DOI: 10.1128/jvi.78.1.158-167.2004
Residues of human cytomegalovirus DNA polymerase catalytic subunit UL54 that are necessary and sufficient for interaction with the accessory protein UL44
Abstract
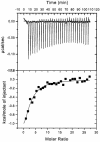
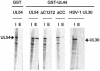
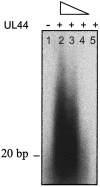
The human cytomegalovirus DNA polymerase contains a catalytic subunit, UL54, and an accessory protein, UL44. Recent studies suggested that UL54 might interact via its extreme C terminus with UL44 (A. Loregian, R. Rigatti, M. Murphy, E. Schievano, G. Palu', and H. S. Marsden, J. Virol. 77:8336-8344, 2003). To address this hypothesis, we quantitatively measured the binding of peptides corresponding to the extreme C terminus of UL54 to UL44 by using isothermal titration calorimetry. A peptide corresponding to the last 22 residues of UL54 was sufficient to bind specifically to UL44 in a 1:1 complex with a dissociation constant of ca. 0.7 microM. To define individual residues in this segment that are crucial for interacting with UL44, we engineered a series of mutations in the C-terminal region of UL54. The UL54 mutants were tested for their ability to interact with UL44 by glutathione S-transferase pulldown assays, for basal DNA polymerase activity, and for long-chain DNA synthesis in the presence of UL44. We observed that deletion of the C-terminal segment or substitution of alanine for Leu1227 or Phe1231 in UL54 greatly impaired both the UL54-UL44 interaction in pulldown assays and long-chain DNA synthesis without affecting basal polymerase activity, identifying these residues as important for subunit interaction. Thus, like the herpes simplex virus UL30-UL42 interaction, a few specific side chains in the C terminus of UL54 are crucial for UL54-UL44 interaction. However, the UL54 residues important for interaction with UL44 are hydrophobic and not basic. This information might aid in the rational design of new drugs for the treatment of human cytomegalovirus infection.
Figures





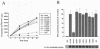
Similar articles
-
Analysis of the association of the human cytomegalovirus DNA polymerase subunit UL44 with the viral DNA replication factor UL84.J Virol. 2009 Aug;83(15):7581-9. doi: 10.1128/JVI.00663-09. Epub 2009 May 20. J Virol. 2009. PMID: 19457994 Free PMC article.
-
Specific residues in the connector loop of the human cytomegalovirus DNA polymerase accessory protein UL44 are crucial for interaction with the UL54 catalytic subunit.J Virol. 2004 Sep;78(17):9084-92. doi: 10.1128/JVI.78.17.9084-9092.2004. J Virol. 2004. PMID: 15308704 Free PMC article.
-
Inhibition of human cytomegalovirus DNA polymerase by C-terminal peptides from the UL54 subunit.J Virol. 2003 Aug;77(15):8336-44. doi: 10.1128/jvi.77.15.8336-8344.2003. J Virol. 2003. PMID: 12857903 Free PMC article.
-
Crystal structure of the cytomegalovirus DNA polymerase subunit UL44 in complex with the C terminus from the catalytic subunit. Differences in structure and function relative to unliganded UL44.J Biol Chem. 2006 Feb 24;281(8):5224-32. doi: 10.1074/jbc.M506900200. Epub 2005 Dec 20. J Biol Chem. 2006. PMID: 16371349
-
Herpesvirus DNA polymerases: Structures, functions and inhibitors.Virus Res. 2017 Apr 15;234:177-192. doi: 10.1016/j.virusres.2017.01.019. Epub 2017 Jan 30. Virus Res. 2017. PMID: 28153606 Review.
Cited by
-
Human cytomegalovirus-induced host protein citrullination is crucial for viral replication.Nat Commun. 2021 Jun 23;12(1):3910. doi: 10.1038/s41467-021-24178-6. Nat Commun. 2021. PMID: 34162877 Free PMC article.
-
The flexible loop of the human cytomegalovirus DNA polymerase processivity factor ppUL44 is required for efficient DNA binding and replication in cells.J Virol. 2009 Sep;83(18):9567-76. doi: 10.1128/JVI.00669-09. Epub 2009 Jul 1. J Virol. 2009. PMID: 19570866 Free PMC article.
-
Association of human cytomegalovirus proteins IRS1 and TRS1 with the viral DNA polymerase accessory subunit UL44.J Gen Virol. 2010 Sep;91(Pt 9):2167-75. doi: 10.1099/vir.0.022640-0. Epub 2010 May 5. J Gen Virol. 2010. PMID: 20444996 Free PMC article.
-
Insights into the Transcriptome of Human Cytomegalovirus: A Comprehensive Review.Viruses. 2023 Aug 8;15(8):1703. doi: 10.3390/v15081703. Viruses. 2023. PMID: 37632045 Free PMC article. Review.
-
Analysis of the association of the human cytomegalovirus DNA polymerase subunit UL44 with the viral DNA replication factor UL84.J Virol. 2009 Aug;83(15):7581-9. doi: 10.1128/JVI.00663-09. Epub 2009 May 20. J Virol. 2009. PMID: 19457994 Free PMC article.
References
-
- Archakov, A. I., V. M. Govorun, A. V. Dubanov, Y. D. Ivanov, A. V. Veselovsky, P. Lewi, and P. Janssen. 2003. Protein-protein interactions as a target for drugs in proteomics. Proteomics 3:380-391. - PubMed
-
- Cihlar, T., M. D. Fuller, and J. M. Cherrington. 1997. Expression of the catalytic subunit (UL54) and the accessory protein (UL44) of human cytomegalovirus DNA polymerase in a coupled in vitro transcription/translation system. Protein Expr. Purif. 11:209-218. - PubMed
-
- Coen, D. M., and P. A. Schaffer. 2003. Antiherpesvirus drugs: a promising spectrum of new drugs and drug targets. Nat. Rev. Drug Discov. 2:278-288. - PubMed
-
- Digard, P., and D. M. Coen. 1990. A novel functional domain of an α-like DNA polymerase. J. Biol. Chem. 265:17393-17396. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

