Evasion of cellular antiviral responses by human cytomegalovirus TRS1 and IRS1
- PMID: 14671101
- PMCID: PMC303427
- DOI: 10.1128/jvi.78.1.197-205.2004
Evasion of cellular antiviral responses by human cytomegalovirus TRS1 and IRS1
Abstract
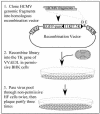
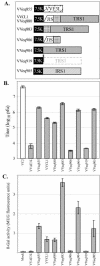
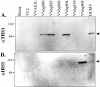
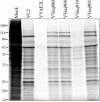
During infection with human cytomegalovirus (HCMV), cellular protein synthesis continues even as viral proteins are being synthesized in abundance. Thus, HCMV may have a mechanism for counteracting host cell antiviral pathways that act by shutting off translation. Consistent with this view, HCMV infection of human fibroblasts rescues the replication of a vaccinia virus mutant lacking the double-stranded RNA-binding protein gene E3L (VVdeltaE3L). HCMV also prevents the phosphorylation of the eukaryotic translation initiation factor eIF-2alpha, the activation of RNase L, and the shutoff of viral and cellular protein synthesis that otherwise result from VVdeltaE3L infection. To identify the HCMV gene(s) responsible for these effects, we prepared a library of VVdeltaE3L recombinants containing HCMV genomic fragments. By infecting nonpermissive cells with this library and screening for VV gene expression and replication, we isolated a virus containing a 2.8-kb HCMV fragment that rescues replication of VVdeltaE3L. The fragment comprises the 3' end of the J1S open reading frame through the entire TRS1 gene. Analyses of additional VVdeltaE3L recombinants revealed that the protein encoded by TRS1, pTRS1, as well as the closely related IRS1 gene, rescues VVdeltaE3L replication and prevent the shutoff of protein synthesis, the phosphorylation of eIF-2alpha, and activation of RNase L. These results demonstrate that TRS1 and IRS1 are able to counteract critical host cell antiviral response pathways.
Figures



References
-
- Abbate, J., J. C. Lacayo, M. Prichard, G. Pari, and M. A. McVoy. 2001. Bifunctional protein conferring enhanced green fluorescence and puromycin resistance. BioTechniques 31:336-340. - PubMed
-
- Biegalke, B. J., and A. P. Geballe. 1990. Translational inhibition by cytomegalovirus transcript leaders. Virology 177:657-667. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

