The ORF2 protein of hepatitis E virus binds the 5' region of viral RNA
- PMID: 14671114
- PMCID: PMC303377
- DOI: 10.1128/jvi.78.1.320-328.2004
The ORF2 protein of hepatitis E virus binds the 5' region of viral RNA
Abstract
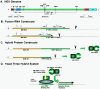
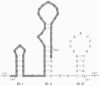
Hepatitis E virus (HEV) is a major human pathogen in much of the developing world. It is a plus-strand RNA virus with a 7.2-kb polyadenylated genome consisting of three open reading frames, ORF1, ORF2, and ORF3. Of these, ORF2 encodes the major capsid protein of the virus and ORF3 encodes a small protein of unknown function. Using the yeast three-hybrid system and traditional biochemical techniques, we have studied the RNA binding activities of ORF2 and ORF3, two proteins encoded in the 3' structural part of the genome. Since the genomic RNA from HEV has been postulated to contain secondary structures at the 5' and 3' ends, we used these two terminal regions, besides other regions within the genome, in this study. Experiments were designed to test for interactions between the genomic RNA fusion constructs with ORF2 and ORF3 hybrid proteins in a yeast cellular environment. We show here that the ORF2 protein contains RNA binding activity. The ORF2 protein specifically bound the 5' end of the HEV genome. Deletion analysis of this protein showed that its RNA binding activity was lost when deletions were made beyond the N-terminal 111 amino acids. Finer mapping of the interacting RNA revealed that a 76-nucleotide (nt) region at the 5' end of the HEV genome was responsible for binding the ORF2 protein. This 76-nt region included the 51-nt HEV sequence, conserved across alphaviruses. Our results support the requirement of this conserved sequence for interaction with ORF2 and also indicate an increase in the strength of the RNA-protein interaction when an additional 44 bases downstream of this 76-nt region were included. Secondary-structure predictions and the location of the ORF2 binding region within the HEV genome indicate that this interaction may play a role in viral encapsidation.
Figures




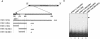
References
-
- Berke, T., and D. O. Matson. 2000. Reclassification of the Calciviridae into distinct genera and exclusion of hepatitis E virus from the family on the basis of comparative phylogenetic analysis. Arch. Virol. 145:1421-1436. - PubMed
-
- Bradley, D. W. 1990. Enterically-transmitted non-A, non-B hepatitis. Br. Med. Bull. 46:442-461. - PubMed
-
- Bradley, D. W., and M. A. Purdy. 1994. Molecular and serological characteristics of hepatitis E virus, p 42-45. In K. Nishioka, H. Suzuki, S. Mishiro, et al. (ed.) Viral hepatitis and liver disease. Springer-Verlag, Tokyo, Japan.
-
- Guarente, L. 1983. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 101:181-191. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

