Characterization of the interaction between P143 and LEF-3 from two different baculovirus species: Choristoneura fumiferana nucleopolyhedrovirus LEF-3 can complement Autographa californica nucleopolyhedrovirus LEF-3 in supporting DNA replication
- PMID: 14671115
- PMCID: PMC303401
- DOI: 10.1128/jvi.78.1.329-339.2004
Characterization of the interaction between P143 and LEF-3 from two different baculovirus species: Choristoneura fumiferana nucleopolyhedrovirus LEF-3 can complement Autographa californica nucleopolyhedrovirus LEF-3 in supporting DNA replication
Abstract
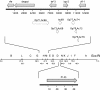
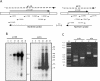
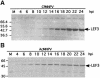
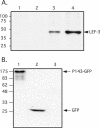
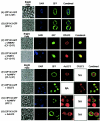
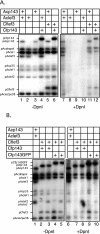
The baculovirus protein P143 is essential for viral DNA replication in vivo, likely as a DNA helicase. We have demonstrated that another viral protein, LEF-3, first described as a single-stranded DNA binding protein, is required for transporting P143 into the nuclei of insect cells. Both of these proteins, along with several other early viral proteins, are also essential for DNA replication in transient assays. We now describe the identification, nucleotide sequences, and transcription patterns of the Choristoneura fumiferana nucleopolyhedrovirus (CfMNPV) homologues of p143 and lef-3 and demonstrate that CfMNPV LEF-3 is also responsible for P143 localization to the nucleus. We predicted that the interaction between P143 and LEF-3 might be critical for cross-species complementation of DNA replication. Support for this hypothesis was generated by substitution of heterologous P143 and LEF-3 between two different baculovirus species, Autographa californica nucleopolyhedrovirus and CfMNPV, in transient DNA replication assays. The results suggest that the P143-LEF-3 complex is an important baculovirus replication factor.
Figures



References
-
- Ahrens, C. H., C. Carlson, and G. F. Rohrmann. 1995. Identification, sequence, and transcriptional analysis of lef-3, a gene essential for Orgyia pseudotsugata baculovirus DNA replication. Virology 210:372-382. - PubMed
-
- Ahrens, C. H., and G. F. Rohrmann. 1996. The DNA polymerase and helicase genes of a baculovirus of Orgyia pseudotsugata. J. Gen. Virol. 77:825-837. - PubMed
-
- Argaud, O., L. Croizier, M. López-Ferber, and G. Croizier. 1998. Two key mutations in the host-range specificity domain of the p143 gene of Autographa californica nucleopolyhedrovirus are required to kill Bombyx mori larvae. J. Gen. Virol. 79:931-935. - PubMed
-
- Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. - PubMed
-
- Bideshi, D. K., and B. A. Federici. 2000. The Trichoplusia ni granulovirus helicase is unable to support replication of Autographa californica multicapsid nucleopolyhedrovirus in cells and larvae of T. ni. J. Gen. Virol. 81:1593-1599. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

