Chromosome-protein interactions in polyomavirus virions
- PMID: 14671132
- PMCID: PMC303386
- DOI: 10.1128/jvi.78.1.513-519.2004
Chromosome-protein interactions in polyomavirus virions
Abstract
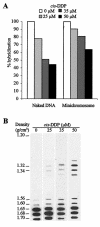
In this work, we sought to determine whether the components of the murine polyomavirus capsid establish specific interactions with the minichromosome encapsidated into the mature viral particles by using the cis-diamminedichloroplatinum(II) cross-linking reagent. Our data indicated that VP1, but not minor capsid proteins, interacts with the viral genome in vivo. In addition, semiquantitative PCR assays performed on cross-linked DNA complexes revealed that VP1 binds to all regions of the viral genome but significantly more to the regulatory region. The implications of such an interaction for viral infectivity are discussed.
Figures

Similar articles
-
VP1, the major capsid protein of the mouse polyomavirus, binds microtubules, promotes their acetylation and blocks the host cell cycle.FEBS J. 2017 Jan;284(2):301-323. doi: 10.1111/febs.13977. Epub 2017 Jan 9. FEBS J. 2017. PMID: 27885808
-
Nuclear localization of avian polyomavirus structural protein VP1 is a prerequisite for the formation of virus-like particles.J Virol. 2004 Jan;78(2):930-7. doi: 10.1128/jvi.78.2.930-937.2004. J Virol. 2004. PMID: 14694124 Free PMC article.
-
Possible role for cellular karyopherins in regulating polyomavirus and papillomavirus capsid assembly.J Virol. 2008 Oct;82(20):9848-57. doi: 10.1128/JVI.01221-08. Epub 2008 Aug 13. J Virol. 2008. PMID: 18701594 Free PMC article.
-
Immunotherapeutic polyoma and human papilloma virus-like particles.Immunotherapy. 2009 Mar;1(2):303-12. doi: 10.2217/1750743X.1.2.303. Immunotherapy. 2009. PMID: 20635947 Review.
-
Microtubules in Polyomavirus Infection.Viruses. 2020 Jan 18;12(1):121. doi: 10.3390/v12010121. Viruses. 2020. PMID: 31963741 Free PMC article. Review.
Cited by
-
Immune sensing of mouse polyomavirus DNA by p204 and cGAS DNA sensors.FEBS J. 2021 Oct;288(20):5964-5985. doi: 10.1111/febs.15962. Epub 2021 May 26. FEBS J. 2021. PMID: 33969628 Free PMC article.
-
A polyomavirus peptide binds to the capsid VP1 pore and has potent antiviral activity against BK and JC polyomaviruses.Elife. 2020 Jan 21;9:e50722. doi: 10.7554/eLife.50722. Elife. 2020. PMID: 31960795 Free PMC article.
-
How viruses use the endoplasmic reticulum for entry, replication, and assembly.Cold Spring Harb Perspect Biol. 2013 Jan 1;5(1):a013250. doi: 10.1101/cshperspect.a013250. Cold Spring Harb Perspect Biol. 2013. PMID: 23284050 Free PMC article. Review.
References
-
- Blasquez, V., A. Stein, C. Ambrose, and M. Bina. 1986. Simian virus 40 protein VP1 is involved in spacing nucleosomes in minichromosomes. J. Mol. Biol. 191:97-106. - PubMed
-
- Brabec, V., and Z. Balcarova. 1993. Restriction-enzyme cleavage of DNA modified by platinum(II) complexes. Eur. J. Biochem. 216:183-187. - PubMed
-
- Buckler-White, A. J., G. W. Humphrey, and V. Pigiet. 1980. Association of polyoma T antigen and DNA with the nuclear matrix from lytically infected 3T6 cells. Cell 22:37-46. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

