p57(Kip2) cooperates with Nurr1 in developing dopamine cells
- PMID: 14671317
- PMCID: PMC307617
- DOI: 10.1073/pnas.2635658100
p57(Kip2) cooperates with Nurr1 in developing dopamine cells
Abstract
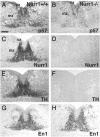
Cyclin-dependent kinase inhibitors of the Cip/Kip family play critical roles in regulating cell proliferation during embryogenesis. However, these proteins also influence cell differentiation by mechanisms that have remained unknown. Here we show that p57Kip2 is expressed in postmitotic differentiating midbrain dopamine cells. Induction of p57Kip2 expression depends on Nurr1, an orphan nuclear receptor that is essential for dopamine neuron development. Moreover, analyses of p57Kip2 gene-targeted mice revealed that p57Kip2 is required for the maturation of midbrain dopamine neuronal cells. Additional experiments in a dopaminergic cell line demonstrated that p57Kip2 can promote maturation by a mechanism that does not require p57Kip2-mediated inhibition of cyclin-dependent kinases. Instead, evidence indicates that p57Kip2 functions by a direct protein-protein interaction with Nurr1. Thus, in addition to its established function in control of proliferation, these results reveal a mechanism whereby p57Kip2 influences postmitotic differentiation of dopamine neurons.
Figures


References
-
- Chellappan, S. P., Giordano, A. & Fisher, P. B. (1998) Curr. Top. Microbiol. Immunol. 227, 57–103. - PubMed
-
- Vidal, A. & Koff, A. (2000) Gene 247, 1–15. - PubMed
-
- Cunningham, J. J. & Roussel, M. F. (2001) Cell Growth Differ. 12, 387–396. - PubMed
-
- Yan, Y., Frisen, J., Lee, M. H., Massague, J. & Barbacid, M. (1997) Genes Dev. 11, 973–983. - PubMed
-
- Zhang, P., Liegeois, N. J., Wong, C., Finegold, M., Hou, H., Thompson, J. C., Silverman, A., Harper, J. W., DePinho, R. A. & Elledge, S. J. (1997) Nature 387, 151–158. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

