Limited boundaries for extensive horizontal gene transfer among Salmonella pathogens
- PMID: 14671318
- PMCID: PMC307627
- DOI: 10.1073/pnas.2634406100
Limited boundaries for extensive horizontal gene transfer among Salmonella pathogens
Abstract
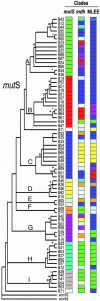
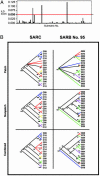
Recombination is thought to be rare within Salmonella, as evidenced by absence of gene transfer among SARC strains that represent the broad genetic diversity of the eight primary subspecies of this common facultative intracellular pathogen. We adopted a phylogenetic approach to assess recombination within the mutS gene of 70 SARB strains, a genetically homogeneous population of Salmonella enterica subspecies I strains, which have in common the ability to infect warm-blooded animals. We report here that SARB strains show evidence for widespread recombinational exchange in contrast to results obtained with strains exhibiting species-level genetic variation. Besides extensive allele shuffling, SARB strains showed notably larger recombinagenic patch sizes for mutS (at least approximately 1.1 kb) than previously reported for S. enterica SARC strains. Explaining these experimental dichotomies provides important insight for understanding microbial evolution, because they suggest likely ecologic and genetic barriers that limit extensive gene transfer in the feral setting.
Figures

References
-
- Perna, N. T., Plunkett, G., III, Burland, V., Mau, B., Glasner, J. D., Rose, D. J., Mayhew, G. F., Evans, P. S., Gregor, J., Kirkpatrick, H. A., et al. (2001) Nature 409, 529–533. - PubMed
-
- Brown, E. W., Kotewicz, M. L. & Cebula, T. A. (2002) Mol. Phylogenet. Evol. 24, 102–120. - PubMed
-
- Denamur, E., Lecointre, G., Darlu, P., Tenaillon, O., Acquaviva, C., Sayada, C., Sunjevaric, I., Rothstein, R., Elion, J., Taddei, F., et al. (2000) Cell 103, 711–721. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

