Crystal structures that suggest late development of genetic code components for differentiating aromatic side chains
- PMID: 14671330
- PMCID: PMC307575
- DOI: 10.1073/pnas.2136794100
Crystal structures that suggest late development of genetic code components for differentiating aromatic side chains
Abstract
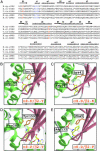
Early forms of the genetic code likely generated "statistical" proteins, with similar side chains occupying the same sequence positions at different ratios. In this scenario, groups of related side chains were treated by aminoacyl-tRNA synthetases as a single molecular species until a discrimination mechanism developed that could separate them. The aromatic amino acids tryptophan, tyrosine, and phenylalanine likely constituted one of these groups. A crystal structure of human tryptophanyl-tRNA synthetase was solved at 2.1 A with a tryptophanyl-adenylate bound at the active site. A cocrystal structure of an active fragment of human tyrosyl-tRNA synthetase with its cognate amino acid analog was also solved at 1.6 A. The two structures enabled active site identifications and provided the information for structure-based sequence alignments of approximately 45 orthologs of each enzyme. Two critical positions shared by all tyrosyl-tRNA synthetases and tryptophanyl-tRNA synthetases for amino acid discrimination were identified. The variations at these two positions and phylogenetic analyses based on the structural information suggest that, in contrast to many other amino acids, discrimination of tyrosine from tryptophan occurred late in the development of the genetic code.
Figures



Similar articles
-
Tryptophanyl-tRNA synthetase crystal structure reveals an unexpected homology to tyrosyl-tRNA synthetase.Structure. 1995 Jan 15;3(1):17-31. doi: 10.1016/s0969-2126(01)00132-0. Structure. 1995. PMID: 7743129
-
Crystal structure of human tryptophanyl-tRNA synthetase catalytic fragment: insights into substrate recognition, tRNA binding, and angiogenesis activity.J Biol Chem. 2004 Feb 27;279(9):8378-88. doi: 10.1074/jbc.M311284200. Epub 2003 Dec 5. J Biol Chem. 2004. PMID: 14660560
-
Crystal structure of Pyrococcus horikoshii tryptophanyl-tRNA synthetase and structure-based phylogenetic analysis suggest an archaeal origin of tryptophanyl-tRNA synthetase.Nucleic Acids Res. 2010 Mar;38(4):1401-12. doi: 10.1093/nar/gkp1053. Epub 2009 Nov 26. Nucleic Acids Res. 2010. PMID: 19942682 Free PMC article.
-
Substrate selection by aminoacyl-tRNA synthetases.Nucleic Acids Symp Ser. 1995;(33):40-2. Nucleic Acids Symp Ser. 1995. PMID: 8643392 Review.
-
Structure, function and evolution of seryl-tRNA synthetases: implications for the evolution of aminoacyl-tRNA synthetases and the genetic code.J Mol Evol. 1995 May;40(5):519-30. doi: 10.1007/BF00166620. J Mol Evol. 1995. PMID: 7540217 Review.
Cited by
-
An asymmetric structure of bacterial TrpRS supports the half-of-the-sites catalytic mechanism and facilitates antimicrobial screening.Nucleic Acids Res. 2023 May 22;51(9):4637-4649. doi: 10.1093/nar/gkad278. Nucleic Acids Res. 2023. PMID: 37070195 Free PMC article.
-
Selective Inhibition of Bacterial Tryptophanyl-tRNA Synthetases by Indolmycin Is Mechanism-based.J Biol Chem. 2016 Jan 1;291(1):255-65. doi: 10.1074/jbc.M115.690321. Epub 2015 Nov 9. J Biol Chem. 2016. PMID: 26555258 Free PMC article.
-
Structure of Leishmania major methionyl-tRNA synthetase in complex with intermediate products methionyladenylate and pyrophosphate.Biochimie. 2011 Mar;93(3):570-82. doi: 10.1016/j.biochi.2010.11.015. Epub 2010 Dec 7. Biochimie. 2011. PMID: 21144880 Free PMC article.
-
The structure of tryptophanyl-tRNA synthetase from Giardia lamblia reveals divergence from eukaryotic homologs.J Struct Biol. 2010 Aug;171(2):238-43. doi: 10.1016/j.jsb.2010.04.010. Epub 2010 May 8. J Struct Biol. 2010. PMID: 20438846 Free PMC article.
-
Structural basis for recognition of cognate tRNA by tyrosyl-tRNA synthetase from three kingdoms.Nucleic Acids Res. 2007;35(13):4289-300. doi: 10.1093/nar/gkm417. Epub 2007 Jun 18. Nucleic Acids Res. 2007. PMID: 17576676 Free PMC article.
References
-
- Ibba, M., Morgan, S., Curnow, A. W., Pridmore, D. R., Vothknecht, U. C., Gardner, W., Lin, W., Woese, C. R. & Söll, D. (1997) Science 278, 1119–1122. - PubMed
-
- Carter, C. W., Jr. (1993) Annu. Rev. Biochem. 62, 715–748. - PubMed
-
- Cavarelli, J. & Moras, D. (1993) FASEB J. 7, 79–86. - PubMed
-
- Cusack, S. (1997) Curr. Opin. Struct. Biol. 7, 881–889. - PubMed
-
- Ribas de Pouplana, L. & Schimmel, P. (2001) Cell 104, 191–193. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

