Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli
- PMID: 14672541
- PMCID: PMC317293
- DOI: 10.1186/1471-2199-4-11
Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli
Abstract
Background: The lambda Red recombineering technology has been used extensively in Escherichia coli and Salmonella typhimurium for easy PCR-mediated generation of deletion mutants, but less so in pathogenic species of E. coli such as EHEC and EPEC. Our early experiments with the use of lambda Red in EHEC and EPEC have led to sporadic results, leading to the present study to identify factors that might improve the efficiency of Red recombineering in these pathogenic strains of E. coli.
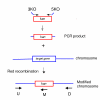
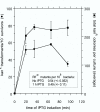
Results: In this report, we have identified conditions that optimize the use of lambda Red for recombineering in EHEC and EPEC. Using plasmids that contain a Ptac-red-gam operon and a temperature-sensitive origin of replication, we have generated multiple mutations (both marked and unmarked) in known virulence genes. In addition, we have easily deleted five O157-specific islands (O-islands) of EHEC suspected of containing virulence factors. We have examined the use of both PCR-generated substrates (40 bp of flanking homology) and plasmid-derived substrates (approximately 1 kb of flanking homology); both work well and each have their own advantages. The establishment of the hyper-rec phenotype requires only a 20 minute IPTG induction period of red and gam. This recombinogenic window is important as constitutive expression of red and gam induces a 10-fold increase in spontaneous resistance to rifampicin. Other factors such as the orientation of the drug marker in recombination substrates and heat shock effects also play roles in the success of Red-mediated recombination in EHEC and EPEC.
Conclusions: The lambda Red recombineering technology has been optimized for use in pathogenic species of E. coli, namely EHEC and EPEC. As demonstration of this technology, five O-islands of EHEC were easily and precisely deleted from the chromosome by electroporation with PCR-generated substrates containing drug markers flanked with 40 bp of target DNA. These results should encourage the use of lambda Red recombineering in these and other strains of pathogenic bacteria for faster identification of virulence factors and the speedy generation of bacterial mutants for vaccine development.
Figures
Similar articles
-
Rapid allelic exchange in enterohemorrhagic Escherichia coli (EHEC) and other E. coli using lambda red recombination.Curr Protoc Microbiol. 2006 Jan;Chapter 5:Unit5A.2. doi: 10.1002/9780471729259.mc05a02s00. Curr Protoc Microbiol. 2006. PMID: 18770591
-
Detection of toxB, a plasmid virulence gene of Escherichia coli O157, in enterohemorrhagic and enteropathogenic E. coli.J Clin Microbiol. 2005 Aug;43(8):4052-6. doi: 10.1128/JCM.43.8.4052-4056.2005. J Clin Microbiol. 2005. PMID: 16081950 Free PMC article.
-
Construction and functional characterization of an integrative form lambda Red recombineering Escherichia coli strain.FEMS Microbiol Lett. 2010 Aug 1;309(2):178-83. doi: 10.1111/j.1574-6968.2010.02036.x. Epub 2010 Jun 16. FEMS Microbiol Lett. 2010. PMID: 20618864
-
[Recombineering and its application].Yi Chuan Xue Bao. 2003 Oct;30(10):983-8. Yi Chuan Xue Bao. 2003. PMID: 14669518 Review. Chinese.
-
λ Recombination and Recombineering.EcoSal Plus. 2016 May;7(1):10.1128/ecosalplus.ESP-0011-2015. doi: 10.1128/ecosalplus.ESP-0011-2015. EcoSal Plus. 2016. PMID: 27223821 Free PMC article. Review.
Cited by
-
Serine Deamination Is a New Acid Tolerance Mechanism Observed in Uropathogenic Escherichia coli.mBio. 2022 Dec 20;13(6):e0296322. doi: 10.1128/mbio.02963-22. Epub 2022 Dec 5. mBio. 2022. PMID: 36468870 Free PMC article.
-
The NleE/OspZ family of effector proteins is required for polymorphonuclear transepithelial migration, a characteristic shared by enteropathogenic Escherichia coli and Shigella flexneri infections.Infect Immun. 2008 Jan;76(1):369-79. doi: 10.1128/IAI.00684-07. Epub 2007 Nov 5. Infect Immun. 2008. PMID: 17984206 Free PMC article.
-
The BarA-UvrY two-component system regulates virulence in avian pathogenic Escherichia coli O78:K80:H9.Infect Immun. 2006 Aug;74(8):4900-9. doi: 10.1128/IAI.00412-06. Infect Immun. 2006. PMID: 16861679 Free PMC article.
-
A recombineering based approach for high-throughput conditional knockout targeting vector construction.Nucleic Acids Res. 2007;35(8):e64. doi: 10.1093/nar/gkm163. Epub 2007 Apr 10. Nucleic Acids Res. 2007. PMID: 17426124 Free PMC article.
-
AI-3 synthesis is not dependent on luxS in Escherichia coli.J Bacteriol. 2006 Aug;188(16):5668-81. doi: 10.1128/JB.00648-06. J Bacteriol. 2006. PMID: 16885435 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

