CONREAL: conserved regulatory elements anchored alignment algorithm for identification of transcription factor binding sites by phylogenetic footprinting
- PMID: 14672977
- PMCID: PMC314294
- DOI: 10.1101/gr.1642804
CONREAL: conserved regulatory elements anchored alignment algorithm for identification of transcription factor binding sites by phylogenetic footprinting
Abstract
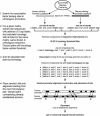
Prediction of transcription-factor target sites in promoters remains difficult due to the short length and degeneracy of the target sequences. Although the use of orthologous sequences and phylogenetic footprinting approaches may help in the recognition of conserved and potentially functional sequences, correct alignment of the short transcription-factor binding sites can be problematic for established algorithms, especially when aligning more divergent species. Here, we report a novel phylogenetic footprinting approach, CONREAL, that uses biologically relevant information, that is, potential transcription-factor binding sites as represented by positional weight matrices, to establish anchors between orthologous sequences and to guide promoter sequence alignment. Comparison of the performance of CONREAL with the global alignment programs LAGAN and AVID using a reference data set, shows that CONREAL performs equally well for closely related species like rodents and human, and has a clear added value for aligning promoter elements of more divergent species like human and fish, as it identifies conserved transcription-factor binding sites that are not found by other methods. CONREAL is accessible via a Web interface at http://conreal.niob.knaw.nl/.
Figures
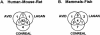


References
-
- Boffelli, D., McAuliffe, J., Ovcharenko, D., Lewis, K.D., Ovcharenko, I., Pachter, L., and Rubin, E.M. 2003. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science 299: 1391-1394. - PubMed
WEB SITE REFERENCES
-
- http://conreal.niob.knaw.nl/; CONREAL Web server.
-
- http://www.emboss.org; EMBOSS package.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
