1-Mb resolution array-based comparative genomic hybridization using a BAC clone set optimized for cancer gene analysis
- PMID: 14672980
- PMCID: PMC314295
- DOI: 10.1101/gr.1847304
1-Mb resolution array-based comparative genomic hybridization using a BAC clone set optimized for cancer gene analysis
Abstract
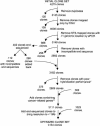
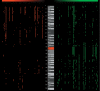
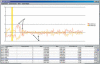
Array-based comparative genomic hybridization (aCGH) is a recently developed tool for genome-wide determination of DNA copy number alterations. This technology has tremendous potential for disease-gene discovery in cancer and developmental disorders as well as numerous other applications. However, widespread utilization of a CGH has been limited by the lack of well characterized, high-resolution clone sets optimized for consistent performance in aCGH assays and specifically designed analytic software. We have assembled a set of approximately 4100 publicly available human bacterial artificial chromosome (BAC) clones evenly spaced at approximately 1-Mb resolution across the genome, which includes direct coverage of approximately 400 known cancer genes. This aCGH-optimized clone set was compiled from five existing sets, experimentally refined, and supplemented for higher resolution and enhancing mapping capabilities. This clone set is associated with a public online resource containing detailed clone mapping data, protocols for the construction and use of arrays, and a suite of analytical software tools designed specifically for aCGH analysis. These resources should greatly facilitate the use of aCGH in gene discovery.
Figures

References
-
- Fritz, B., Schubert, F., Wrobel, G., Schwaenen, C., Wessendorf, S., Nessling, M., Korz, C., Rieker, R.J., Montgomery, K., Kucherlapati, R., et al. 2002. Microarray-based copy number and expression profiling in dedifferentiated and pleomorphic liposarcoma. Cancer Res. 62: 2993-2998. - PubMed
-
- Hodgson, G., Hager, J.H., Volik, S., Hariono, S., Wernick, M., Moore, D., Nowak, N., Albertson, D.G., Pinkel, D., Collins, C., et al. 2001. Genome scanning with array CGH delineates regional alterations in mouse islet carcinomas. Nat. Genet. 29: 459-464. - PubMed
WEB SITE REFERENCES
-
- http://www.ncbi.nlm.nih.gov/genome/sts/epcr.cgi; National Center for Biotechnology Information ePCR search.
-
- http://www.genome.ucsc.edu/; Human Genome Sequence browser.
-
- http://www.tigr.org/tdb/at/abe/bac_end_search.html; TIGR human BAC clone resource.
-
- http://acgh.afcri.upenn.edu; Weber Lab aCGH resource site containing all clone data and analytic software.
-
- http://www.tigr.org/software/tm4/mev.html; TIGR Multi Experiment Viewer software.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
