Transcriptome analysis of chlamydial growth during IFN-gamma-mediated persistence and reactivation
- PMID: 14673075
- PMCID: PMC307677
- DOI: 10.1073/pnas.2535394100
Transcriptome analysis of chlamydial growth during IFN-gamma-mediated persistence and reactivation
Abstract
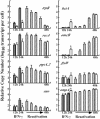
Chlamydia trachomatis is an obligatory intracellular prokaryotic parasite that causes a spectrum of clinically important chronic inflammatory diseases of humans. Persistent infection may play a role in the pathophysiology of chlamydial disease. Here we describe the chlamydial transcriptome in an in vitro model of IFN-gamma-mediated persistence and reactivation from persistence. Tryptophan utilization, DNA repair and recombination, phospholipid utilization, protein translation, and general stress genes were up-regulated during persistence. Down-regulated genes included chlamydial late genes and genes involved in proteolysis, peptide transport, and cell division. Persistence was characterized by altered but active biosynthetic processes and continued replication of the chromosome. On removal of IFN-gamma, chlamydiae rapidly reentered the normal developmental cycle and reversed transcriptional changes associated with cytokine treatment. The coordinated transcriptional response to IFN-gamma implies that a chlamydial response stimulon has evolved to control the transition between acute and persistent growth of the pathogen. In contrast to the paradigm of persistence as a general stress response, our findings suggest that persistence is an alternative life cycle used by chlamydiae to avoid the host immune response.
Figures


References
-
- Schachter, J. (1978) N. Engl. J. Med. 298, 428-434. - PubMed
-
- Morrison, R. P., Manning, D. S. & Caldwell, H. D. (1992) in Advances in Host Defense Mechanisms, ed. Quinn, T. C. (Raven, New York), pp. 57-84.
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

