Specific and covalent targeting of conjugating and deconjugating enzymes of ubiquitin-like proteins
- PMID: 14673145
- PMCID: PMC303361
- DOI: 10.1128/MCB.24.1.84-95.2004
Specific and covalent targeting of conjugating and deconjugating enzymes of ubiquitin-like proteins
Abstract
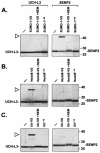
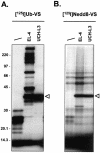
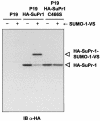
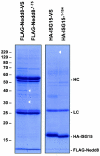
Modification of proteins by ubiquitin (Ub)-like proteins (UBLs) plays an important role in many cellular processes, including cell cycle progression, nuclear transport, and autophagy. Protein modification occurs via UBL-conjugating and -deconjugating enzymes, which presumably exert a regulatory function by determining the conjugation status of the substrate proteins. To target and identify UBL-modifying enzymes, we produced Nedd8, ISG15, and SUMO-1 in Escherichia coli and equipped them with a C-terminal electrophilic trap (vinyl sulfone [VS]) via an intein-based method. These C-terminally modified UBL probes reacted with purified UBL-activating (E1), -conjugating (E2), and -deconjugating enzymes in a covalent fashion. Modified UBLs were radioiodinated and incubated with cell lysates prepared from mouse cell lines and tissues to allow visualization of polypeptides reactive with individual UBL probes. The cell type- and tissue-specific labeling patterns observed for the UBL probes reflect distinct expression profiles of active enzymes, indicating tissue-specific functions of UBLs. We identify Ub C-terminal hydrolase L1 (UCH-L1) and DEN1/NEDP1/SENP8, in addition to UCH-L3, as proteases with specificity for Nedd8. The Ub-specific protease isopeptidase T/USP5 is shown to react with ISG15-VS. Furthermore, we demonstrate that the desumoylation enzyme SuPr-1 can be modified by SUMO-1-VS, a modification that is dependent on the SuPr-1 active-site cysteine. The UBL probes described here will be valuable tools for the further characterization of the enzymatic pathways that govern modification by UBLs.
Figures




References
-
- Bayer, P., A. Arndt, S. Metzger, R. Mahajan, F. Melchior, R. Jaenicke, and J. Becker. 1998. Structure determination of the small ubiquitin-related modifier SUMO-1. J. Mol. Biol. 280:275-286. - PubMed
-
- Bernier-Villamor, V., D. A. Sampson, M. J. Matunis, and C. D. Lima. 2002. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108:345-356. - PubMed
-
- Best, J. L., S. Ganiatsas, S. Agarwal, A. Changou, P. Salomoni, O. Shirihai, P. B. Meluh, P. P. Pandolfi, and L. I. Zon. 2002. SUMO-1 protease-1 regulates gene transcription through PML. Mol. Cell 10:843-855. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
