Dual effects of IkappaB kinase beta-mediated phosphorylation on p105 Fate: SCF(beta-TrCP)-dependent degradation and SCF(beta-TrCP)-independent processing
- PMID: 14673179
- PMCID: PMC303339
- DOI: 10.1128/MCB.24.1.475-486.2004
Dual effects of IkappaB kinase beta-mediated phosphorylation on p105 Fate: SCF(beta-TrCP)-dependent degradation and SCF(beta-TrCP)-independent processing
Abstract
Processing of the p105 NF-kappaB precursor to yield the p50 active subunit is a unique and rare case in which the ubiquitin system is involved in limited processing rather than in complete destruction of its target. The mechanisms involved in this process are largely unknown, although a glycine repeat in the middle of p105 has been identified as a processing stop signal. IkappaB kinase (IKK)beta-mediated phosphorylation at the C-terminal domain with subsequent recruitment of the SCF(beta-TrCP) ubiquitin ligase leads to accelerated processing and degradation of the precursor, yet the roles that the kinase and ligase play in each of these two processes have not been elucidated. Here we demonstrate that IKKbeta has two distinct functions: (i) stimulation of degradation and (ii) stimulation of processing. IKKbeta-induced degradation is dependent on SCF(beta-TrCP), which acts through multiple lysine residues in the IkappaBgamma domain. In contrast, IKKbeta-induced processing of p105 is beta-transduction repeat-containing protein (beta-TrCP) independent, as it is not affected by expression of a dominant-negative beta-TrCP or following its silencing by small inhibitory RNA. Furthermore, removal of all 30 lysine residues from IkappaBgamma results in complete inhibition of IKK-dependent degradation but has no effect on IKK-dependent processing. Yet processing still requires the activity of the ubiquitin system, as it is inhibited by dominant-negative UbcH5a. We suggest that IKKbeta mediates its two distinct effects by affecting, directly and indirectly, two different E3s.
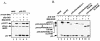
Figures







References
-
- Baldi, L., K. Brown, G. Franzoso, and U. Siebenlist. 1996. Critical role for lysines 21 and 22 in signal-induced, ubiquitin-mediated proteolysis of IκBα. J. Biol. Chem. 271:376-379. - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
-
- Cohen, S., A. Orian, and A. Ciechanover. 2001. Processing of p105 is inhibited by docking of p50 active subunits to the ankyrin repeat domain, and inhibition is alleviated by signaling via the carboxyl-terminal phosphorylation/ubiquitin-ligase binding domain. J. Biol. Chem. 276:26769-26776. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
