Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes
- PMID: 14676319
- PMCID: PMC307626
- DOI: 10.1073/pnas.2532165100
Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes
Abstract
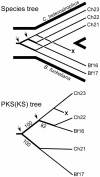
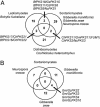
Fungal type I polyketides (PKs) are synthesized by PK synthases (PKSs) and include well known secondary metabolites such as the anticholesterol drug lovastatin and the potent natural carcinogen aflatoxin. Other type I PKs are known to be virulence factors for some plant pathogens and pigments such as melanin. In this study, a phylogenomic approach was used to investigate the origin and diversity of fungal genes encoding putative PKSs that are predicted to synthesize type I PKs. The resulting genealogy, constructed by using the highly conserved PKS ketosynthase (KS) domain, indicated that: (i). Species within subphylum Pezizomycotina (phylum Ascomycota) but not early diverging ascomycetes, like Saccharomyces cerevisiae (Saccharomycotina) or Schizosaccharomyces pombe (Taphrinomycotina), had large numbers (7-25) of PKS genes. (ii). Bacteria and fungi had separate groups of PKS genes; the few exceptions are the likely result of horizontal gene transfer from bacteria to various sublineages of fungi. (iii). The bulk of genes encoding fungal PKSs fell into eight groups. Four groups were predicted to synthesize variously reduced PKs, and four groups were predicted to make unreduced PKs. (iv). Species within different classes of Pezizomycotina shared the same groups of PKS genes. (v). Different fungal genomes shared few putative orthologous PKS genes, even between closely related genomes in the same class or genus. (vi) The discontinuous distributions of orthologous PKSs among fungal species can be explained by gene duplication, divergence, and gene loss; horizontal gene transfer among fungi does not need to be invoked.
Figures

References
-
- Hopwood, D. A. (1997) Chem. Rev. 97, 2465–2497. - PubMed
-
- Hendrickson, L., Davis, C. R., Roach, C., Nguyen, D. K., McAda, P. C. & Reeves, C. D. (1999) Chem. Biol. 6, 429–439. - PubMed
-
- Abe, Y, Suzuki, T., Mizvno, T., Ono, C., Iwamoto, K., Hosobuchi, M. & Yoshikawa, H. (2002) Mol. Genet. Genomics 267, 636–646. - PubMed
-
- Berbee, M. L. (2001) Physiol. Mol. Plant Pathol. 59, 165–187.
-
- Galagan, J. E., Calvo, S. E., Borkovich, K. A., Selker, E. U., Read, N. D., Jaffe, D., FitzHugh, W., Ma, L. J., Smirnov, S., Purcell, S., et al. (2003) Nature 442, 859–868. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

