The Genomic Threading Database: a comprehensive resource for structural annotations of the genomes from key organisms
- PMID: 14681393
- PMCID: PMC308777
- DOI: 10.1093/nar/gkh043
The Genomic Threading Database: a comprehensive resource for structural annotations of the genomes from key organisms
Abstract
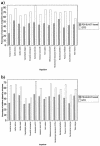
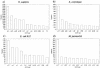
Currently, the Genomic Threading Database (GTD) contains structural assignments for the proteins encoded within the genomes of nine eukaryotes and 101 prokaryotes. Structural annotations are carried out using a modified version of GenTHREADER, a reliable fold recognition method. The Gen THREADER annotation jobs are distributed across multiple clusters of processors using grid technology and the predictions are deposited in a relational database accessible via a web interface at http://bioinf.cs.ucl.ac.uk/GTD. Using this system, up to 84% of proteins encoded within a genome can be confidently assigned to known folds with 72% of the residues aligned. On average in the GTD, 64% of proteins encoded within a genome are confidently assigned to known folds and 58% of the residues are aligned to structures.
Figures
References
-
- Hoersch S., Leroy,C., Brown,N.P. andrade,M.A. and Sander,C. (2000) The GeneQuiz web server: protein functional analysis through the Web. Trends Biochem. Sci., 25, 33–35. - PubMed
-
- Bryson K., Luck,M., Joy,M. and Jones,D.T. (2000) Applying agents to bioinformatics in GeneWeaver. Lect. Notes Artif. Int., 1860, 60–71.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

