GeneDB: a resource for prokaryotic and eukaryotic organisms
- PMID: 14681429
- PMCID: PMC308742
- DOI: 10.1093/nar/gkh007
GeneDB: a resource for prokaryotic and eukaryotic organisms
Abstract
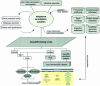
GeneDB (http://www.genedb.org/) is a genome database for prokaryotic and eukaryotic organisms. The resource provides a portal through which data generated by the Pathogen Sequencing Unit at the Wellcome Trust Sanger Institute and other collaborating sequencing centres can be made publicly available. It combines data from finished and ongoing genome and expressed sequence tag (EST) projects with curated annotation, that can be searched, sorted and downloaded, using a single web based resource. The current release stores 11 datasets of which six are curated and maintained by biologists, who review and incorporate information from the scientific literature, public databases and the respective research communities.
Figures

References
-
- Rice P., Longden,I. and Bleasby,A. (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet., 16, 276–277. - PubMed
-
- Nielsen H. and Krogh,A. (1998) Prediction of signal peptides and signal anchors by a hidden Markov model. Proc. Int. Conf. Intell. Syst. Mol. Biol., 6, 122–130. - PubMed
-
- Sonnhammer E.L., von Heijne,G. and Krogh,A. (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol., 6, 175–182. - PubMed
-
- Rutherford K., Parkhill,J., Crook,J., Horsnell,T., Rice,P., Rajandream,M.A. and Barrell,B. (2000) Artemis: sequence visualization and annotation. Bioinformatics, 16, 944–945. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

