The Mouse Genome Database (MGD): integrating biology with the genome
- PMID: 14681461
- PMCID: PMC308859
- DOI: 10.1093/nar/gkh125
The Mouse Genome Database (MGD): integrating biology with the genome
Abstract
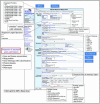
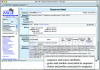
The Mouse Genome Database (MGD) is one component of the Mouse Genome Informatics (MGI) system (http://www.informatics.jax.org), a community database resource for the laboratory mouse. MGD strives to provide a comprehensive knowledgebase about the mouse with experiments and data annotated from both literature and online sources. MGD curates and presents consensus and experimental data representations of genetic, genotype (sequence) and phenotype information including highly detailed reports about genes and gene products. Primary foci of integration are through representations of relationships between genes, sequences and phenotypes. MGD collaborates with other bioinformatics groups to curate a definitive set of information about the laboratory mouse and to build and implement the data and semantic standards that are essential for comparative genome analysis. Recent developments in MGD discussed here include an extensive integration of the mouse sequence data and substantial revisions in the presentation, query and visualization of sequence data.
Figures


References
-
- FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I & II Team (2002) Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature, 420, 563–573. - PubMed
-
- Strausberg R.L., Feingold,E.A., Klausner,R.D. and Collins, F.S. (1999) The mammalian gene collection. Science, 286, 455–457. - PubMed

