Human protein reference database as a discovery resource for proteomics
- PMID: 14681466
- PMCID: PMC308804
- DOI: 10.1093/nar/gkh070
Human protein reference database as a discovery resource for proteomics
Abstract
The rapid pace at which genomic and proteomic data is being generated necessitates the development of tools and resources for managing data that allow integration of information from disparate sources. The Human Protein Reference Database (http://www.hprd.org) is a web-based resource based on open source technologies for protein information about several aspects of human proteins including protein-protein interactions, post-translational modifications, enzyme-substrate relationships and disease associations. This information was derived manually by a critical reading of the published literature by expert biologists and through bioinformatics analyses of the protein sequence. This database will assist in biomedical discoveries by serving as a resource of genomic and proteomic information and providing an integrated view of sequence, structure, function and protein networks in health and disease.
Figures

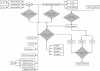


References
-
- The International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. - PubMed
-
- Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. - PubMed
-
- Mann M. and Pandey,A. (2001) Use of mass spectrometry-derived data to annotate nucleotide and protein sequence databases. Trends Biochem. Sci., 26, 54–61. - PubMed
-
- Tucker C.L., Gera,J.F. and Uetz,P. (2001) Towards an understanding of complex protein networks. Trends Cell Biol., 11, 102–116. - PubMed
-
- Peri S., Navarro,J.D., Amanchy,R., Kristiansen,T.Z., Jonnalagadda,C.K., Surendranath,V., Niranjan,V., Muthusamy,B., Gandhi,T.K.B., Gronborg,M. et.al. (2003) Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res., 13, 2363–2371. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

