Mutation of CERKL, a novel human ceramide kinase gene, causes autosomal recessive retinitis pigmentosa (RP26)
- PMID: 14681825
- PMCID: PMC1181900
- DOI: 10.1086/381055
Mutation of CERKL, a novel human ceramide kinase gene, causes autosomal recessive retinitis pigmentosa (RP26)
Abstract
Retinitis pigmentosa (RP), the main cause of adult blindness, is a genetically heterogeneous disorder characterized by progressive loss of photoreceptors through apoptosis. Up to now, 39 genes and loci have been implicated in nonsyndromic RP, yet the genetic bases of >50% of the cases, particularly of the recessive forms, remain unknown. Previous linkage analysis in a Spanish consanguineous family allowed us to define a novel autosomal recessive RP (arRP) locus, RP26, within an 11-cM interval (17.4 Mb) on 2q31.2-q32.3. In the present study, we further refine the RP26 locus down to 2.5 Mb, by microsatellite and single-nucleotide polymorphism (SNP) homozygosity mapping. After unsuccessful mutational analysis of the nine genes initially reported in this region, a detailed gene search based on expressed-sequence-tag data was undertaken. We finally identified a novel gene encoding a ceramide kinase (CERKL), which encompassed 13 exons. All of the patients from the RP26 family bear a homozygous mutation in exon 5, which generates a premature termination codon. The same mutation was also characterized in another, unrelated, Spanish pedigree with arRP. Human CERKL is expressed in the retina, among other adult and fetal tissues. A more detailed analysis by in situ hybridization on adult murine retina sections shows expression of Cerkl in the ganglion cell layer. Ceramide kinases convert the sphingolipid metabolite ceramide into ceramide-1-phosphate, both key mediators of cellular apoptosis and survival. Ceramide metabolism plays an essential role in the viability of neuronal cells, the membranes of which are particularly rich in sphingolipids. Therefore, CERKL deficiency could shift the relative levels of the signaling sphingolipid metabolites and increase sensitivity of photoreceptor and other retinal cells to apoptotic stimuli. This is the first genetic report suggesting a direct link between retinal neurodegeneration in RP and sphingolipid-mediated apoptosis.
Figures




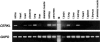

References
Electronic-Database Information
-
- BCM Search Launcher, http://searchlauncher.bcm.tmc.edu/
-
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/ (for the new SNPs observed in the present study, which are deposited under the following identification numbers: ss12676100, ss12676101, ss12676102, ss12676103, ss12676104, ss12676105, ss12676106, ss12676107, ss12676108, ss12676109, ss12676110, ss12676111, ss12676112, ss12676113, and ss12676114)
-
- EMBnet BoxShade Server, http://www.ch.embnet.org/software/BOX_form.html
-
- GenBank, http://www.ncbi.nih.gov/Genbank/ (for truncated EST [accession number BE797822], partial cDNA [accession number BC020465], human SPHK1 [accession number AAF73423], murine SPHK1 [accession number AAC61697], human SPHK2 [accession number AAF74124], murine SPHK2 [accession number AAF74125], Drosophila SPHK [accession number AAF48045], C. intestinalis SPHK [accession number AK112588], yeast Lcb4p [accession number NP_014814], yeast Lcb5p [NP_013361], A. thaliana SPHK [accession number AY128394], human CERK [accession number AB079066], murine CERK [accession number AB079067], C. intestinalis CERK [accession number AK112750], Drosophila CERK [accession number AAF52040], human CERKL [accession number AY357073], murine CERKL [accession numbers BY742285 and BC046474], C. elegans CERK [accession number AAC67466], human DGKb [accession number Q9Y6T7], murine DGKb [accession number XP_147651], human DGKe [accession number NP_003638], murine DGKe [accession number NP_062378], C. elegans DGK1 [accession number NP_508190], A. thaliana DGK1 [accession number Q39017], Drosophila DGK2 [accession number Q09103], and Drosophila DGK1 [accession number Q01583])
-
- NCBI BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
References
-
- Bayés M, Goldaracena B, Martínez-Mir A, Iragui-Madoz MI, Solans T, Chivelet P, Bussaglia E, Ramos-Arroyo MA, Baiget M, Vilageliu L, Balcells S, González-Duarte R, Grinberg D (1998) A new autosomal recessive retinitis pigmentosa locus maps on chromosome 2q31-q33. J Med Genet 35:141–145 - PMC - PubMed
-
- Bayés M, Martínez-Mir A, Valverde D, del Rio E, Vilageliu L, Grinberg D, Balcells S, Ayuso C, Baiget M, Gonzàlez-Duarte R (1996) Autosomal recessive retinitis pigmentosa in Spain: evaluation of four genes and two loci involved in the disease. Clin Genet 50:380–387 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

