Population-genetic basis of haplotype blocks in the 5q31 region
- PMID: 14681827
- PMCID: PMC1181911
- DOI: 10.1086/381040
Population-genetic basis of haplotype blocks in the 5q31 region
Abstract
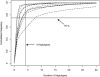
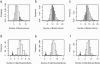
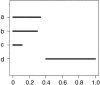
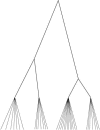
We investigated patterns of nucleotide variation in the 5q31 region identified by Daly et al. as containing haplotype blocks, to determine whether the blocklike pattern requires the assumption of hotspots in recombination. Using extensive simulations that generate data matched to the Daly et al. data set in (a) the method of ascertainment of single-nucleotide polymorphisms, (b) the heterozygosity of ascertained markers, (c) the number of block boundaries, and (d) the diversity of haplotypes within blocks, we show that the patterns found in the Daly et al. data are not consistent with the assumption of uniform recombination in a population of constant size but are consistent either with the presence of hotspots in a population of constant size or with the absence of hotspots if there was a period of rapid population growth. We further show that estimates of local recombination rate can distinguish between population growth and hotspots as the primary cause of a blocklike pattern. Estimates of local recombination rates for the Daly et al. data do not indicate the presence of recombination hotspots.
Figures



References
Electronic-Database Information
-
- makesamples, http://home.uchicago.edu/~rhudson1/source/mksamples.html (for the program for generating samples under neutral models)
-
- maxhap, http://home.uchicago.edu/~rhudson1/source/maxhap.html (for the program for estimating ρ (4Nr), a crossing-over parameter, and f(=g/r), a gene conversion parameter, through use of a maximum composite likelihood method)
-
- MDBlocks, http://ib.berkeley.edu/labs/slatkin/eriq/software/mdb_web/index.htm (for the haplotype block–partitioning program)
-
- Whitehead/MIT Center for Genome Research, http://www-genome.wi.mit.edu/humgen/IBD5/index.html (for 5q31 data)
References
-
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D (2002) The structure of haplotype blocks in the human genome. Science 296:2225–222910.1126/science.1069424 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

