A post genomic characterization of Arabidopsis ferredoxins
- PMID: 14684843
- PMCID: PMC316305
- DOI: 10.1104/pp.103.032755
A post genomic characterization of Arabidopsis ferredoxins
Abstract
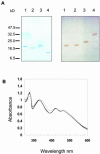
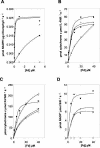
In higher plant plastids, ferredoxin (Fd) is the unique soluble electron carrier protein located in the stroma. Consequently, a wide variety of essential metabolic and signaling processes depend upon reduction by Fd. The currently available plant genomes of Arabidopsis and rice (Oryza sativa) contain several genes encoding putative Fds, although little is known about the proteins themselves. To establish whether this variety represents redundancy or specialized function, we have recombinantly expressed and purified the four conventional [2Fe-2S] Fd proteins encoded in the Arabidopsis genome and analyzed their physical and functional properties. Two proteins are leaf type Fds, having relatively low redox potentials and supporting a higher photosynthetic activity. One protein is a root type Fd, being more efficiently reduced under nonphotosynthetic conditions and supporting a higher activity of sulfite reduction. A further Fd has a remarkably positive redox potential and so, although redox active, is limited in redox partners to which it can donate electrons. Immunological analysis indicates that all four proteins are expressed in mature leaves. This holistic view demonstrates how varied and essential soluble electron transfer functions in higher plants are fulfilled through a diversity of Fd proteins.
Figures




References
-
- Akashi T, Matsumura T, Ideguchi T, Iwakiri K, Kawakatsu T, Taniguchi I, Hase T (1999) Comparison of the electrostatic binding sites on the surface of ferredoxin for two ferredoxin-dependent enzymes, ferredoxin-NADP(+) reductase and sulfite reductase. J Biol Chem 274: 29399-29405 - PubMed
-
- Aliverti A, Faber R, Finnerty CM, Ferioli C, Pandini V, Negri A, Karplus PA, Zanetti G (2001) Biochemical and crystallographic characterization of ferredoxin-NADP(+) reductase from nonphotosynthetic tissues. Biochemistry 40: 14501-14508 - PubMed
-
- Aliverti A, Hagen WR, Zanetti G (1995) Direct electrochemistry and EPR spectroscopy of spinach ferredoxin mutants with modified electron transfer properties. FEBS Lett 368: 220-224 - PubMed
-
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

