migS, a cis-acting site that affects bipolar positioning of oriC on the Escherichia coli chromosome
- PMID: 14685268
- PMCID: PMC1271666
- DOI: 10.1038/sj.emboj.7600028
migS, a cis-acting site that affects bipolar positioning of oriC on the Escherichia coli chromosome
Abstract
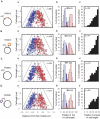
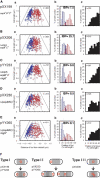
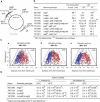
During replication of the Escherichia coli chromosome, the replicated Ori domains migrate towards opposite cell poles, suggesting that a cis-acting site for bipolar migration is located in this region. To identify this cis-acting site, a series of mutants was constructed by splitting subchromosomes from the original chromosome. One mutant, containing a 720 kb subchromosome, was found to be defective in the bipolar positioning of oriC. The creation of deletion mutants allowed the identification of migS, a 25 bp sequence, as the cis-acting site for the bipolar positioning of oriC. When migS was located at the replication terminus, the chromosomal segment showed bipolar positioning. migS was able to rescue bipolar migration of plasmid DNA containing a mutation in the SopABC partitioning system. Interestingly, multiple copies of the migS sequence on a plasmid in trans inhibited the bipolar positioning of oriC. Taken together, these findings indicate that migS plays a crucial role in the bipolar positioning of oriC. In addition, real-time analysis of the dynamic morphological changes of nucleoids in wild-type and migS mutants suggests that bipolar positioning of the replicated oriC contributes to nucleoid organization.
Figures




References
-
- Ben-Yehuda S, Rudner DZ, Losick R (2003) RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299: 532–536 - PubMed
-
- Draper GC, Gober JW (2002) Bacterial chromosome segregation. Annu Rev Microbiol 56: 567–597 - PubMed
-
- Edwards DH, Errington J (1997) The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol Microbiol 24: 905–915 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

