Identification of a redox-regulated chaperone network
- PMID: 14685279
- PMCID: PMC1271656
- DOI: 10.1038/sj.emboj.7600016
Identification of a redox-regulated chaperone network
Abstract
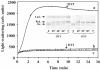
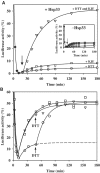
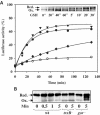
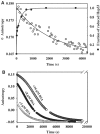
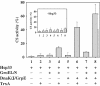
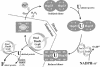
We have identified and reconstituted a multicomponent redox-chaperone network that appears to be designed to protect proteins against stress-induced unfolding and to refold proteins when conditions return to normal. The central player is Hsp33, a redox-regulated molecular chaperone. Hsp33, which is activated by disulfide bond formation and subsequent dimerization, works as an efficient chaperone holdase that binds to unfolding protein intermediates and maintains them in a folding competent conformation. Reduction of Hsp33 is catalyzed by the glutaredoxin and thioredoxin systems in vivo, and leads to the formation of highly active, reduced Hsp33 dimers. Reduction of Hsp33 is necessary but not sufficient for substrate protein release. Substrate dissociation from Hsp33 is linked to the presence of the DnaK/DnaJ/GrpE foldase system, which alone, or in concert with the GroEL/GroES system, then supports the refolding of the substrate proteins. Upon substrate release, reduced Hsp33 dimers dissociate into inactive monomers. This regulated substrate transfer ultimately links substrate release and Hsp33 inactivation to the presence of available DnaK/DnaJ/GrpE, and, therefore, to the return of cells to non-stress conditions.
Figures
References
-
- Barbirz S, Jakob U, Glocker MO (2000) Mass spectrometry unravels disulfide bond formation as the mechanism that activates a molecular chaperone. J Biol Chem 275: 18759–18766 - PubMed
-
- Beissinger M, Buchner J (1998) How chaperones fold proteins. Biol Chem 379: 245–259 - PubMed
-
- Buchberger A, Schroder H, Buttner M, Valencia A, Bukau B (1994) A conserved loop in the ATPase domain of the DnaK chaperone is essential for stable binding of GrpE. Nat Struct Biol 1: 95–101 - PubMed
-
- Buchner J, Schmidt M, Fuchs M, Jaenicke R, Rudolph R, Schmid FX, Kiefhaber T (1991) GroE facilitates refolding of citrate synthase by suppressing aggregation. Biochemistry 30: 1586–1591 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

