Role of the C-terminal lysine residues of streptococcal surface enolase in Glu- and Lys-plasminogen-binding activities of group A streptococci
- PMID: 14688086
- PMCID: PMC343989
- DOI: 10.1128/IAI.72.1.94-105.2004
Role of the C-terminal lysine residues of streptococcal surface enolase in Glu- and Lys-plasminogen-binding activities of group A streptococci
Abstract
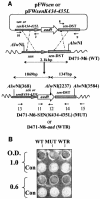
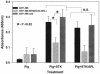
Streptococcal surface enolase (SEN) is a major plasminogen-binding protein of group A streptococci. Our earlier biochemical studies have suggested that the region responsible for this property is likely located at the C-terminal end of the SEN molecule. In the present study, the gene encoding SEN was cloned from group A streptococci M6 isolate D471. A series of mutations in the sen gene corresponding to the C-terminal region (428KSFYNLKK435) of the SEN molecule were created by either deleting one or more terminal lysine residues or replacing them with leucine. All purified recombinant SEN proteins with altered C-terminal ends were found to be enzymatically active and were analyzed for their Glu- and Lys-plasminogen-binding activities. Wild-type SEN bound to Lys-plasminogen with almost three times more affinity than to Glu-plasminogen. However, the recombinant mutant SEN proteins with a deletion of Lys434-435 or with K435L and K434-435L replacements showed a significant decrease in Glu- and Lys-plasminogen-binding activities. Accordingly, a streptococcal mutant expressing SEN-K434-435L showed a significant decrease in Glu- and Lys-plasminogen-binding activities. Biochemical and functional analyses of the isogenic mutant strain revealed a significant decrease in its abilities to cleave a chromogenic tripeptide substrate, acquire plasminogen from human plasma, and penetrate the extracellular matrix. Together, these data indicate that the last two C-terminal lysine residues of surface-exposed SEN contribute significantly to the plasminogen-binding activity of intact group A streptococci and hence to their ability to exploit host properties to their own advantage in tissue invasion.
Figures

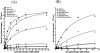


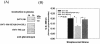
Similar articles
-
Defining the structural basis of human plasminogen binding by streptococcal surface enolase.J Biol Chem. 2009 Jun 19;284(25):17129-17137. doi: 10.1074/jbc.M109.004317. Epub 2009 Apr 10. J Biol Chem. 2009. PMID: 19363026 Free PMC article.
-
alpha-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci.J Biol Chem. 1998 Jun 5;273(23):14503-15. doi: 10.1074/jbc.273.23.14503. J Biol Chem. 1998. PMID: 9603964
-
Plasminogen-mediated group A streptococcal adherence to and pericellular invasion of human pharyngeal cells.Microb Pathog. 2003 Dec;35(6):293-303. doi: 10.1016/j.micpath.2003.08.004. Microb Pathog. 2003. PMID: 14580393
-
High-Resolution Single-Particle Cryo-EM Hydrated Structure of Streptococcus pyogenes Enolase Offers Insights into Its Function as a Plasminogen Receptor.Biochemistry. 2023 Feb 7;62(3):735-746. doi: 10.1021/acs.biochem.2c00637. Epub 2023 Jan 26. Biochemistry. 2023. PMID: 36701429
-
Group A Streptococcus exploits human plasminogen for bacterial translocation across epithelial barrier via tricellular tight junctions.Sci Rep. 2016 Jan 29;7:20069. doi: 10.1038/srep20069. Sci Rep. 2016. PMID: 26822058 Free PMC article.
Cited by
-
An octamer of enolase from Streptococcus suis.Protein Cell. 2012 Oct;3(10):769-80. doi: 10.1007/s13238-012-2040-7. Epub 2012 Oct 11. Protein Cell. 2012. PMID: 23055041 Free PMC article.
-
Relationships Between Plasminogen-Binding M-Protein and Surface Enolase for Human Plasminogen Acquisition and Activation in Streptococcus pyogenes.Front Microbiol. 2022 May 24;13:905670. doi: 10.3389/fmicb.2022.905670. eCollection 2022. Front Microbiol. 2022. PMID: 35685926 Free PMC article.
-
Potential of known and short prokaryotic protein motifs as a basis for novel peptide-based antibacterial therapeutics: a computational survey.Front Microbiol. 2014 Jan 21;5:4. doi: 10.3389/fmicb.2014.00004. eCollection 2014. Front Microbiol. 2014. PMID: 24478765 Free PMC article.
-
The interaction of streptococcal enolase with canine plasminogen: the role of surfaces in complex formation.PLoS One. 2014 Feb 10;9(2):e88395. doi: 10.1371/journal.pone.0088395. eCollection 2014. PLoS One. 2014. PMID: 24520380 Free PMC article.
-
Identification of plasminogen-binding sites in Streptococcus suis enolase that contribute to bacterial translocation across the blood-brain barrier.Front Cell Infect Microbiol. 2024 Feb 22;14:1356628. doi: 10.3389/fcimb.2024.1356628. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38456079 Free PMC article.
References
-
- Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M.-Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. M. Leung, P. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. - PMC - PubMed
-
- Berge, A., and U. Sjobring. 1993. PAM, a novel plasminogen-binding protein from Streptococcus pyogenes. J. Biol. Chem. 268:25417-25424. - PubMed
-
- Bergmann, S., M. Rohde, G. S. Chhatwal, and S. Hammerschmidt. 2001. α-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 40:1273-1287. - PubMed
-
- Bergmann, S., D. Wild, O. Diekmann, R. Frank, D. Bracht, G. S. Chhatwal, and S. Hammerschmidt. 2003. Identification of a novel plasmin(ogen)-binding motif in surface displayed α-enolase of Streptococcus pneumoniae. Mol. Microbiol. 49:411-423. - PubMed
-
- Bishop, C., D. M. Rankine, and J. H. Talbott. 1959. The nucleotide in normal human blood. J. Biol. Chem. 234:1233-1237. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

