Cross-sectional analysis of clinical and environmental isolates of Pseudomonas aeruginosa: biofilm formation, virulence, and genome diversity
- PMID: 14688090
- PMCID: PMC343948
- DOI: 10.1128/IAI.72.1.133-144.2004
Cross-sectional analysis of clinical and environmental isolates of Pseudomonas aeruginosa: biofilm formation, virulence, and genome diversity
Abstract
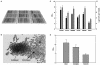
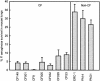
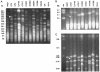
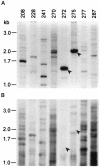
Chronic lung infections with Pseudomonas aeruginosa biofilms are associated with refractory and fatal pneumonia in cystic fibrosis (CF). In this study, a group of genomically diverse P. aeruginosa isolates were compared with the reference strain PAO1 to assess the roles of motility, twitching, growth rate, and overproduction of a capsular polysaccharide (alginate) in biofilm formation. In an in vitro biofilm assay system, P. aeruginosa displayed strain-specific biofilm formation that was not solely dependent on these parameters. Compared with non-CF isolates, CF isolates expressed two opposing growth modes: reduced planktonic growth versus efficient biofilm formation. Planktonic cells of CF isolates showed elevated sensitivity to hydrogen peroxide, a reactive oxygen intermediate, and decreased lung colonization in an aerosol infection mouse model. Despite having identical genomic profiles, CF sequential isolates produced different amounts of biofilm. While P. aeruginosa isolates exhibited genomic diversity, the genome size of these isolates was estimated to be 0.4 to 19% (27 to 1,184 kb) larger than that of PAO1. To identify these extra genetic materials, random amplification of polymorphic DNA was coupled with PAO1-subtractive hybridization. Three loci were found within the genomes of two CF isolates encoding one novel homolog involved in retaining a Shigella virulence plasmid (mvpTA) and two divergent genes that function in removing negative supercoiling (topA) and biosynthesis of pyoverdine (PA2402). Together, P. aeruginosa biodiversity could provide one cause for the variation of morbidity and mortality in CF. P. aeruginosa may possess undefined biofilm adhesins that are important to the development of an antibiofilm therapeutic target.
Figures


Similar articles
-
Pseudomonas aeruginosa cystic fibrosis isolates of similar RAPD genotype exhibit diversity in biofilm forming ability in vitro.BMC Microbiol. 2010 Feb 8;10:38. doi: 10.1186/1471-2180-10-38. BMC Microbiol. 2010. PMID: 20141637 Free PMC article.
-
Genotypic and Phenotypic Diversity of Staphylococcus aureus Isolates from Cystic Fibrosis Patient Lung Infections and Their Interactions with Pseudomonas aeruginosa.mBio. 2020 Jun 23;11(3):e00735-20. doi: 10.1128/mBio.00735-20. mBio. 2020. PMID: 32576671 Free PMC article.
-
Anaerobic culture conditions favor biofilm-like phenotypes in Pseudomonas aeruginosa isolates from patients with cystic fibrosis.FEMS Immunol Med Microbiol. 2006 Dec;48(3):373-80. doi: 10.1111/j.1574-695X.2006.00157.x. Epub 2006 Oct 18. FEMS Immunol Med Microbiol. 2006. PMID: 17052266
-
Persistent infections and immunity in cystic fibrosis.Front Biosci. 2002 Feb 1;7:d442-57. doi: 10.2741/a787. Front Biosci. 2002. PMID: 11815305 Review.
-
Microevolution of Pseudomonas aeruginosa to a chronic pathogen of the cystic fibrosis lung.Curr Top Microbiol Immunol. 2013;358:91-118. doi: 10.1007/82_2011_199. Curr Top Microbiol Immunol. 2013. PMID: 22311171 Review.
Cited by
-
Targeting bacterial topoisomerase I to meet the challenge of finding new antibiotics.Future Med Chem. 2015;7(4):459-71. doi: 10.4155/fmc.14.157. Future Med Chem. 2015. PMID: 25875873 Free PMC article. Review.
-
Antibiofilm effect of biofilm-dispersing agents on clinical isolates of Pseudomonas aeruginosa with various biofilm structures.J Microbiol. 2018 Dec;56(12):902-909. doi: 10.1007/s12275-018-8336-4. Epub 2018 Oct 25. J Microbiol. 2018. PMID: 30361978
-
Swarming motility, secretion of type 3 effectors and biofilm formation phenotypes exhibited within a large cohort of Pseudomonas aeruginosa clinical isolates.J Med Microbiol. 2010 May;59(Pt 5):511-520. doi: 10.1099/jmm.0.017715-0. Epub 2010 Jan 21. J Med Microbiol. 2010. PMID: 20093376 Free PMC article.
-
Pseudomonas aeruginosa cystic fibrosis isolates of similar RAPD genotype exhibit diversity in biofilm forming ability in vitro.BMC Microbiol. 2010 Feb 8;10:38. doi: 10.1186/1471-2180-10-38. BMC Microbiol. 2010. PMID: 20141637 Free PMC article.
-
Bacterial fitness in chronic wounds appears to be mediated by the capacity for high-density growth, not virulence or biofilm functions.PLoS Pathog. 2019 Mar 20;15(3):e1007511. doi: 10.1371/journal.ppat.1007511. eCollection 2019 Mar. PLoS Pathog. 2019. PMID: 30893371 Free PMC article.
References
-
- Alm, R. A., and J. S. Mattick. 1995. Identification of a gene, pilV, required for type 4 fimbrial biogenesis in Pseudomonas aeruginosa, whose product possesses a pre-pilin-like leader sequence. Mol. Microbiol. 16:485-496. - PubMed
-
- Barth, A. L., and T. L. Pitt. 1996. The high amino-acid content of sputum from cystic fibrosis patients promotes growth of auxotrophic Pseudomonas aeruginosa. J. Med. Microbiol. 45:110-119. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

