Legionella pneumophila type II protein secretion promotes virulence in the A/J mouse model of Legionnaires' disease pneumonia
- PMID: 14688110
- PMCID: PMC344012
- DOI: 10.1128/IAI.72.1.310-321.2004
Legionella pneumophila type II protein secretion promotes virulence in the A/J mouse model of Legionnaires' disease pneumonia
Abstract
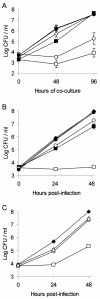
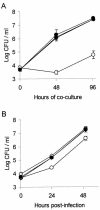
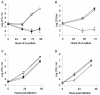
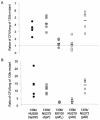
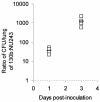
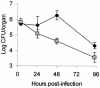
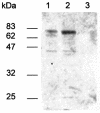
Legionella pneumophila, the gram-negative agent of Legionnaires' disease, possesses type IV pili and a type II protein secretion (Lsp) system, both of which are dependent upon the PilD prepilin peptidase. By analyzing multiple pilD mutants and various types of Lsp mutants as well as performing trans-complementation of these mutants, we have confirmed that PilD and type II secretion genes are required for L. pneumophila infection of both amoebae and human macrophages. Based upon a complete analysis of lspDE, lspF, and lspG mutants, we found that the type II system controls the secretion of protease, RNase, lipase, phospholipase A, phospholipase C, lysophospholipase A, and tartrate-sensitive and tartrate-resistant acid phosphatase activities and influences the appearance of colonies. Examination of the developing L. pneumophila genome database indicated that the organism has two other loci (lspC and lspLM) that are predicted to promote secretion and thus a set of genes that is comparable to the type II secretion genes in other gram-negative bacteria. In contrast to lsp mutants, L. pneumophila pilus mutants lacking either the PilQ secretin, the PspA pseudopilin, or pilin were not defective for colonial growth, secreted activities, or intracellular replication. L. pneumophila dot/icm mutants were also not impaired for type II-dependent exoenzymes. Upon intratracheal inoculation into A/J mice, lspDE, lspF, and pilD mutants, but not pilus mutants, exhibited a reduced ability to grow in the lung, as measured by competition assays. The lspF mutant was also defective in an in vivo kinetic assay. Examination of infected mouse sera revealed that type II secreted proteins are expressed in vivo. Thus, the L. pneumophila Lsp system is a virulence factor and the only type II secretion system linked to intracellular infection.
Figures
Similar articles
-
Type II protein secretion is a subset of the PilD-dependent processes that facilitate intracellular infection by Legionella pneumophila.Infect Immun. 2001 Apr;69(4):2092-8. doi: 10.1128/IAI.69.4.2092-2098.2001. Infect Immun. 2001. PMID: 11254562 Free PMC article.
-
The type II protein secretion system of Legionella pneumophila promotes growth at low temperatures.J Bacteriol. 2004 Jun;186(12):3712-20. doi: 10.1128/JB.186.12.3712-3720.2004. J Bacteriol. 2004. PMID: 15175284 Free PMC article.
-
The prepilin peptidase is required for protein secretion by and the virulence of the intracellular pathogen Legionella pneumophila.Mol Microbiol. 1999 Feb;31(3):959-70. doi: 10.1046/j.1365-2958.1999.01239.x. Mol Microbiol. 1999. PMID: 10048038
-
Pathogenicity of Legionella pneumophila.Int J Med Microbiol. 2001 Nov;291(5):331-43. doi: 10.1078/1438-4221-00139. Int J Med Microbiol. 2001. PMID: 11727817 Review.
-
Secreted phospholipases of the lung pathogen Legionella pneumophila.Int J Med Microbiol. 2018 Jan;308(1):168-175. doi: 10.1016/j.ijmm.2017.10.002. Epub 2017 Oct 28. Int J Med Microbiol. 2018. PMID: 29108710 Review.
Cited by
-
The major facilitator superfamily-type protein LbtC promotes the utilization of the legiobactin siderophore by Legionella pneumophila.Microbiology (Reading). 2012 Mar;158(Pt 3):721-735. doi: 10.1099/mic.0.055533-0. Epub 2011 Dec 8. Microbiology (Reading). 2012. PMID: 22160401 Free PMC article.
-
Mouse macrophages are permissive to motile Legionella species that fail to trigger pyroptosis.Infect Immun. 2010 Jan;78(1):423-32. doi: 10.1128/IAI.00070-09. Epub 2009 Oct 19. Infect Immun. 2010. PMID: 19841075 Free PMC article.
-
Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung.Proc Natl Acad Sci U S A. 2006 Dec 12;103(50):19146-51. doi: 10.1073/pnas.0608279103. Epub 2006 Dec 5. Proc Natl Acad Sci U S A. 2006. PMID: 17148602 Free PMC article.
-
Zinc Metalloprotease ProA from Legionella pneumophila Inhibits the Pro-Inflammatory Host Response by Degradation of Bacterial Flagellin.Biomolecules. 2022 Apr 22;12(5):624. doi: 10.3390/biom12050624. Biomolecules. 2022. PMID: 35625552 Free PMC article.
-
Legionella pneumophila type II secretion dampens the cytokine response of infected macrophages and epithelia.Infect Immun. 2011 May;79(5):1984-97. doi: 10.1128/IAI.01077-10. Epub 2011 Mar 7. Infect Immun. 2011. PMID: 21383054 Free PMC article.
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Aragon, V., O. Rossier, and N. P. Cianciotto. 2002. Legionella pneumophila genes that encode lipase and phospholipase C activities. Microbiology 148:2223-2231. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

