Recognition of dileucine-based sorting signals from HIV-1 Nef and LIMP-II by the AP-1 gamma-sigma1 and AP-3 delta-sigma3 hemicomplexes
- PMID: 14691137
- PMCID: PMC2173715
- DOI: 10.1083/jcb.200307157
Recognition of dileucine-based sorting signals from HIV-1 Nef and LIMP-II by the AP-1 gamma-sigma1 and AP-3 delta-sigma3 hemicomplexes
Abstract
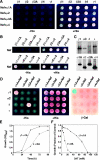
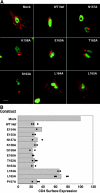
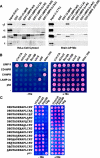
The sorting of transmembrane proteins to endosomes and lysosomes is mediated by signals present in the cytosolic tails of the proteins. A subset of these signals conform to the [DE]XXXL[LI] consensus motif and mediate sorting via interactions with heterotetrameric adaptor protein (AP) complexes. However, the identity of the AP subunits that recognize these signals remains controversial. We have used a yeast three-hybrid assay to demonstrate that [DE]XXXL[LI]-type signals from the human immunodeficiency virus negative factor protein and the lysosomal integral membrane protein II interact with combinations of the gamma and sigma1 subunits of AP-1 and the delta and sigma3 subunits of AP-3, but not the analogous combinations of AP-2 and AP-4 subunits. The sequence requirements for these interactions are similar to those for binding to the whole AP complexes in vitro and for function of the signals in vivo. These observations reveal a novel mode of recognition of sorting signals involving the gamma/delta and sigma subunits of AP-1 and AP-3.
Figures



References
-
- Aguilar, R.C., H. Ohno, K.C. Roche, and J.S. Bonifacino. 1997. Functional domain mapping of the clathrin-associated adaptor medium chains mu1 and mu2. J. Biol. Chem. 272:2760–2766. - PubMed
-
- Aiken, C., J. Konner, N.R. Landau, M.E. Lenburg, and D. Trono. 1994. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell. 76:853–864. - PubMed
-
- Barriocanal, J.G., J.S. Bonifacino, L. Yuan, and I.V. Sandoval. 1986. Biosynthesis, glycosylation, movement through the Golgi system, and transport to lysosomes by an N-linked carbohydrate-independent mechanism of three lysosomal integral membrane proteins. J. Biol. Chem. 261:16755–16763. - PubMed
-
- Bonifacino, J.S., and L.M. Traub. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72:395–447. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

