Active-site alkylation destabilizes human O6-alkylguanine DNA alkyltransferase
- PMID: 14691244
- PMCID: PMC2286524
- DOI: 10.1110/ps.03319404
Active-site alkylation destabilizes human O6-alkylguanine DNA alkyltransferase
Abstract
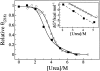
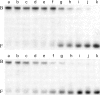
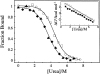
O(6)-alkylguanine-DNA alkyltransferase (AGT) repairs pro-mutagenic O(6)-alkylguanine and O(4)-alkylthymine lesions in DNA. The alkylated form of the protein is not reactivated; instead, it is rapidly ubiquitinated and degraded. Here, we show that alkylation destabilizes the native fold of the protein by 0.5-1.2 kcal/mole and the DNA-binding function by 0.8-1.4 kcal/mole. On this basis, we propose that destabilization of the native conformational ensemble acts as a signal for ubiquitination.
Figures
References
-
- Dalessio, P.M. and Ropson, I.J. 1998. pH dependence of the folding of intestinal fatty acid binding protein. Arch. Biochem. Biophys. 359 199–208. - PubMed
-
- Daniels, D.S. and Tainer, J.A. 2000. Conserved structural motifs governing the stoichiometric repair of alkylated DNA by O6-alkylguanine-DNA alkyltransferase. Mutation Res. 460 151–163. - PubMed
-
- Daniels, D.S., Mol, C.D., Arval, A.S., Kanugula, S., Pegg, A.E., and Tainer, J.A. 2000. Active and alkylated human AGT structures: A novel zinc site, inhibitor and extrahelical binding. DNA damage reversal revealed by mutants and structures of active and alkylated human AGT. EMBO J. 19 1719–1730. - PMC - PubMed
-
- Fried, M.G. 1989. Measurement of protein-DNA interaction parameters by electrophoresis mobility shift assay. Electrophoresis 10 366–376. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

