Molecular basis of intrinsic macrolide resistance in the Mycobacterium tuberculosis complex
- PMID: 14693532
- PMCID: PMC310192
- DOI: 10.1128/AAC.48.1.143-150.2004
Molecular basis of intrinsic macrolide resistance in the Mycobacterium tuberculosis complex
Abstract
The intrinsic resistance of the Mycobacterium tuberculosis complex (MTC) to most antibiotics, including macrolides, is generally attributed to the low permeability of the mycobacterial cell wall. However, nontuberculous mycobacteria (NTM) are much more sensitive to macrolides than members of the MTC. A search for macrolide resistance determinants within the genome of M. tuberculosis revealed the presence of a sequence encoding a putative rRNA methyltransferase. The deduced protein is similar to Erm methyltransferases, which confer macrolide-lincosamide-streptogramin (MLS) resistance by methylation of 23S rRNA, and was named ErmMT. The corresponding gene, ermMT (erm37), is present in all members of the MTC but is absent in NTM species. Part of ermMT is deleted in some vaccine strains of Mycobacterium bovis BCG, such as the Pasteur strain, which lack the RD2 region. The Pasteur strain was susceptible to MLS antibiotics, whereas MTC species harboring the RD2 region were resistant to them. The expression of ermMT in the macrolide-sensitive Mycobacterium smegmatis and BCG Pasteur conferred MLS resistance. The resistance patterns and ribosomal affinity for erythromycin of Mycobacterium host strains expressing ermMT, srmA (monomethyltransferase from Streptomyces ambofaciens), and ermE (dimethyltransferase from Saccharopolyspora erythraea) were compared, and the ones conferred by ErmMT were similar to those conferred by SrmA, corresponding to the MLS type I phenotype. These results suggest that ermMT plays a major role in the intrinsic macrolide resistance of members of the MTC and could be the first example of a gene conferring resistance by target modification in mycobacteria.
Figures


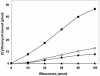
References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. A. Smith, J. G. Seidman, and K. Struhl. 1995. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
-
- Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. - PubMed
-
- Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. - PMC - PubMed
-
- Bussiere, D. E., S. W. Muchmore, C. G. Dealwis, G. Schluckebier, V. L. Nienaber, R. P. Edalji, K. A. Walter, U. S. Ladror, T. F. Holzman, and C. Abad-Zapatero. 1998. Crystal structure of ErmC′, an rRNA methyltransferase which mediates antibiotic resistance in bacteria. Biochemistry 37:7103-7112. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

