Late domain-dependent inhibition of equine infectious anemia virus budding
- PMID: 14694104
- PMCID: PMC368837
- DOI: 10.1128/jvi.78.2.724-732.2004
Late domain-dependent inhibition of equine infectious anemia virus budding
Abstract
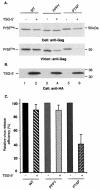
The Gag proteins of a number of different retroviruses contain late or L domains that promote the release of virions from the plasma membrane. Three types of L domains have been identified to date: Pro-Thr-Ala-Pro (PTAP), Pro-Pro-X-Tyr, and Tyr-Pro-Asp-Leu. It has previously been demonstrated that overexpression of the N-terminal, E2-like domain of the endosomal sorting factor TSG101 (TSG-5') inhibits human immunodeficiency virus type 1 (HIV-1) release but does not affect the release of the PPPY-containing retrovirus murine leukemia virus (MLV), whereas overexpression of the C-terminal portion of TSG101 (TSG-3') potently disrupts both HIV-1 and MLV budding. In addition, it has been reported that, while the release of a number of retroviruses is disrupted by proteasome inhibitors, equine infectious anemia virus (EIAV) budding is not affected by these agents. In this study, we tested the ability of TSG-5', TSG-3', and full-length TSG101 (TSG-F) overexpression, a dominant negative form of the AAA ATPase Vps4, and proteasome inhibitors to disrupt the budding of EIAV particles bearing each of the three types of L domain. The results indicate that (i) inhibition by TSG-5' correlates with dependence on PTAP; (ii) the release of wild-type EIAV (EIAV/WT) is insensitive to TSG-3', whereas this C-terminal TSG101 fragment potently impairs the budding of EIAV when it is rendered PTAP or PPPY dependent; (iii) budding of all EIAV clones is blocked by dominant negative Vps4; and (iv) EIAV/WT release is not impaired by proteasome inhibitors, while EIAV/PTAP and EIAV/PPPY release is strongly disrupted by these compounds. These findings highlight intriguing similarities and differences in host factor utilization by retroviral L domains and suggest that the insensitivity of EIAV to proteasome inhibitors is conferred by the L domain itself and not by determinants in Gag outside the L domain.
Figures




Similar articles
-
Defects in human immunodeficiency virus budding and endosomal sorting induced by TSG101 overexpression.J Virol. 2003 Jun;77(11):6507-19. doi: 10.1128/jvi.77.11.6507-6519.2003. J Virol. 2003. PMID: 12743307 Free PMC article.
-
Functional replacement and positional dependence of homologous and heterologous L domains in equine infectious anemia virus replication.J Virol. 2002 Feb;76(4):1569-77. doi: 10.1128/jvi.76.4.1569-1577.2002. J Virol. 2002. PMID: 11799151 Free PMC article.
-
Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function.Proc Natl Acad Sci U S A. 2002 Jan 22;99(2):955-60. doi: 10.1073/pnas.032511899. Proc Natl Acad Sci U S A. 2002. PMID: 11805336 Free PMC article.
-
Retrovirus budding.Virus Res. 2004 Dec;106(2):87-102. doi: 10.1016/j.virusres.2004.08.007. Virus Res. 2004. PMID: 15567490 Review.
-
[Envelope virus assembly and budding].Uirusu. 2010 Jun;60(1):105-13. doi: 10.2222/jsv.60.105. Uirusu. 2010. PMID: 20848870 Review. Japanese.
Cited by
-
Cumulative mutations of ubiquitin acceptor sites in human immunodeficiency virus type 1 gag cause a late budding defect.J Virol. 2006 Jul;80(13):6267-75. doi: 10.1128/JVI.02177-05. J Virol. 2006. PMID: 16775314 Free PMC article.
-
Mechanisms for enveloped virus budding: can some viruses do without an ESCRT?Virology. 2008 Mar 15;372(2):221-32. doi: 10.1016/j.virol.2007.11.008. Epub 2007 Dec 11. Virology. 2008. PMID: 18063004 Free PMC article. Review.
-
Novel approaches to inhibiting HIV-1 replication.Antiviral Res. 2010 Jan;85(1):119-41. doi: 10.1016/j.antiviral.2009.09.009. Epub 2009 Sep 24. Antiviral Res. 2010. PMID: 19782103 Free PMC article. Review.
-
Heterologous late-domain sequences have various abilities to promote budding of human immunodeficiency virus type 1.J Virol. 2005 Jul;79(14):9038-45. doi: 10.1128/jvi.79.14.9038-9045.2005. J Virol. 2005. PMID: 15994797 Free PMC article.
-
YRKL sequence of influenza virus M1 functions as the L domain motif and interacts with VPS28 and Cdc42.J Virol. 2006 Mar;80(5):2291-308. doi: 10.1128/JVI.80.5.2291-2308.2006. J Virol. 2006. Retraction in: J Virol. 2006 Oct;80(20):10289. doi: 10.1128/JVI.01632-06. PMID: 16474136 Free PMC article. Retracted.
References
-
- Amberg, D. C., E. Basart, and D. Botstein. 1995. Defining protein interactions with yeast actin in vivo. Nat. Struct. Biol. 2:28-35. - PubMed
-
- Babst, M., D. J. Katzmann, E. J. Estepa-Sabal, T. Meerloo, and S. D. Emr. 2002. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 3:271-282. - PubMed
-
- Babst, M., D. J. Katzmann, W. B. Snyder, B. Wendland, and S. D. Emr. 2002. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3:283-289. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

